You are browsing environment: FUNGIDB
CAZyme Information: PPTG_17187-t26_1-p1
You are here: Home > Sequence: PPTG_17187-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_17187-t26_1-p1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 44 | 282 | 2.7e-19 | 0.9228295819935691 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 2.73e-13 | 17 | 276 | 39 | 304 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033 | Glyco_hydro_17 | 1.01e-06 | 208 | 283 | 229 | 307 | Glycosyl hydrolases family 17. |
| 237015 | PRK11901 | 8.65e-05 | 290 | 452 | 55 | 240 | hypothetical protein; Reviewed |
| 411384 | Streccoc_I_II | 0.006 | 362 | 454 | 821 | 928 | antigen I/II family LPXTG-anchored adhesin. Members of the antigen I/II family are adhesins with a glucan-binding domain, two types of repetitive regions, an isopeptide bond-forming domain associated with shear resistance, and a C-terminal LPXTG motif for anchoring to the cell wall. They occur in oral Streptococci, and tend to be major cell surface adhesins. Members of this family include SspA and SspB from Streptococcus gordonii, antigen I/II from S. mutans, etc. |
| 223021 | PHA03247 | 0.009 | 353 | 452 | 2719 | 2849 | large tegument protein UL36; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.08e-103 | 9 | 284 | 9 | 288 | |
| 1.80e-92 | 25 | 286 | 574 | 836 | |
| 3.23e-67 | 23 | 284 | 27 | 288 | |
| 5.42e-53 | 23 | 284 | 20 | 282 | |
| 1.12e-42 | 23 | 196 | 27 | 200 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.47e-06 | 34 | 283 | 50 | 292 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
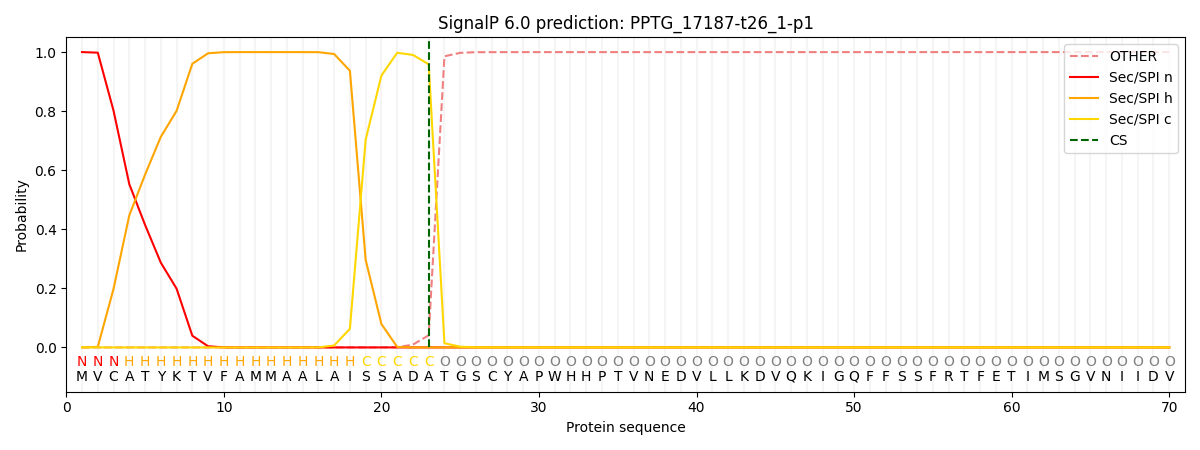
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000229 | 0.999744 | CS pos: 23-24. Pr: 0.9588 |
