You are browsing environment: FUNGIDB
CAZyme Information: PPTG_15600-t26_1-p1
You are here: Home > Sequence: PPTG_15600-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_15600-t26_1-p1 | |||||||||||
| CAZy Family | GT20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:28 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 853 | 1600 | 1.1e-273 | 0.972936400541272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 854 | 1535 | 3 | 701 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 1.15e-27 | 165 | 258 | 6 | 106 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
| 340916 | MFS_GLUT6_8_Class3_like | 1.27e-22 | 1749 | 2161 | 2 | 426 | Glucose transporter (GLUT) types 6 and 8, Class 3 GLUTs, and similar transporters of the Major Facilitator Superfamily. This subfamily is composed of glucose transporter type 6 (GLUT6), GLUT8, plant early dehydration-induced gene ERD6-like proteins, and similar insect proteins including facilitated trehalose transporter Tret1-1. GLUTs, also called Solute carrier family 2, facilitated glucose transporters (SLC2A), are a family of proteins that facilitate the transport of hexoses such as glucose and fructose. There are fourteen GLUTs found in humans; they display different substrate specificities and tissue expression. They have been categorized into three classes based on sequence similarity: Class 1 (GLUTs 1-4, 14); Class 2 (GLUTs 5, 7, 9, and 11); and Class 3 (GLUTs 6, 8, 10, 12, and HMIT). Insect Tret1-1 is a low-capacity facilitative transporter for trehalose that mediates the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source. GLUT proteins are comprised of about 500 amino acid residues, possess a single N-linked oligosaccharide, and have 12 transmembrane segments. They belong to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 395036 | Sugar_tr | 4.81e-18 | 1750 | 2155 | 1 | 428 | Sugar (and other) transporter. |
| 340915 | MFS_GLUT_Class1_2_like | 2.45e-14 | 1755 | 2093 | 6 | 373 | Class 1 and Class 2 Glucose transporters (GLUTs) of the Major Facilitator Superfamily. This subfamily includes Class 1 and Class 2 glucose transporters (GLUTs) including Solute carrier family 2, facilitated glucose transporter member 1 (SLC2A1, also called glucose transporter type 1 or GLUT1), SLC2A2-5 (GLUT2-5), SLC2A7 (GLUT7), SLC2A9 (GLUT9), SLC2A11 (GLUT11), SLC2A14 (GLUT14), and similar proteins. GLUTs are a family of proteins that facilitate the transport of hexoses such as glucose and fructose. There are fourteen GLUTs found in humans; they display different substrate specificities and tissue expression. They have been categorized into three classes based on sequence similarity: Class 1 (GLUTs 1-4, 14); Class 2 (GLUTs 5, 7, 9, and 11); and Class 3 (GLUTs 6, 8, 10, 12, and HMIT). GLUTs 1-5 are the most thoroughly studied and are well-established as glucose and/or fructose transporters in various tissues and cell types. GLUT proteins are comprised of about 500 amino acid residues, possess a single N-linked oligosaccharide, and have 12 transmembrane segments. They belong to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 2245 | 1 | 2246 | |
| 0.0 | 13 | 2235 | 10 | 2241 | |
| 0.0 | 7 | 2239 | 9 | 2225 | |
| 0.0 | 11 | 2235 | 12 | 2231 | |
| 0.0 | 11 | 2241 | 11 | 2225 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.54e-237 | 77 | 1742 | 252 | 1918 | Callose synthase 7 OS=Arabidopsis thaliana OX=3702 GN=CALS7 PE=3 SV=3 |
|
| 1.82e-232 | 83 | 1742 | 253 | 1878 | Callose synthase 9 OS=Arabidopsis thaliana OX=3702 GN=CALS9 PE=2 SV=2 |
|
| 1.11e-227 | 74 | 1742 | 230 | 1934 | Callose synthase 2 OS=Arabidopsis thaliana OX=3702 GN=CALS2 PE=2 SV=3 |
|
| 2.99e-227 | 83 | 1742 | 265 | 1892 | Callose synthase 10 OS=Arabidopsis thaliana OX=3702 GN=CALS10 PE=2 SV=5 |
|
| 5.62e-227 | 74 | 1742 | 230 | 1934 | Callose synthase 1 OS=Arabidopsis thaliana OX=3702 GN=CALS1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000056 | 0.000000 |
TMHMM Annotations download full data without filtering help
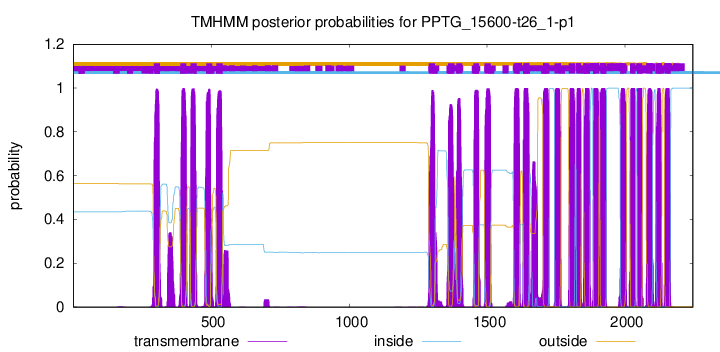
| Start | End |
|---|---|
| 292 | 314 |
| 388 | 410 |
| 425 | 447 |
| 482 | 504 |
| 519 | 541 |
| 1289 | 1311 |
| 1359 | 1381 |
| 1388 | 1410 |
| 1451 | 1470 |
| 1491 | 1513 |
| 1596 | 1618 |
| 1631 | 1653 |
| 1703 | 1725 |
| 1745 | 1767 |
| 1796 | 1818 |
| 1823 | 1840 |
| 1850 | 1872 |
| 1884 | 1906 |
| 1910 | 1932 |
| 1983 | 2005 |
| 2018 | 2037 |
| 2044 | 2062 |
| 2077 | 2099 |
| 2111 | 2133 |
| 2143 | 2165 |
