You are browsing environment: FUNGIDB
CAZyme Information: PPTG_15443-t26_1-p1
You are here: Home > Sequence: PPTG_15443-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
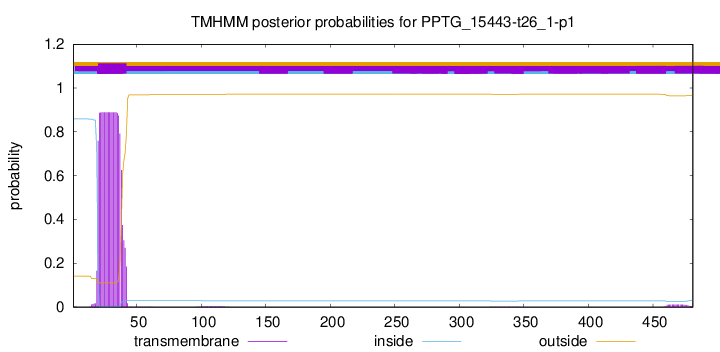
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_15443-t26_1-p1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT71 | 106 | 364 | 3.8e-42 | 0.946969696969697 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 402574 | Mannosyl_trans3 | 9.53e-25 | 108 | 338 | 10 | 242 | Mannosyltransferase putative. This family is conserved in fungi. Several members are annotated as being alpha-1,3-mannosyltransferase but this could not be confirmed. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.01e-122 | 103 | 479 | 62 | 432 | |
| 4.78e-113 | 96 | 478 | 128 | 516 | |
| 2.43e-92 | 100 | 388 | 66 | 358 | |
| 3.68e-65 | 21 | 462 | 11 | 453 | |
| 3.33e-58 | 100 | 450 | 65 | 437 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.37e-09 | 100 | 405 | 358 | 683 | Putative alpha-1,3-mannosyltransferase MNN1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN1 PE=2 SV=1 |
|
| 1.84e-08 | 117 | 386 | 215 | 482 | Probable alpha-1,3-mannosyltransferase MNT4 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT4 PE=3 SV=1 |
|
| 7.24e-08 | 97 | 386 | 172 | 460 | Alpha-1,3-mannosyltransferase MNT2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNT2 PE=1 SV=1 |
|
| 2.37e-06 | 169 | 331 | 423 | 578 | Putative alpha-1,3-mannosyltransferase MNN13 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN13 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
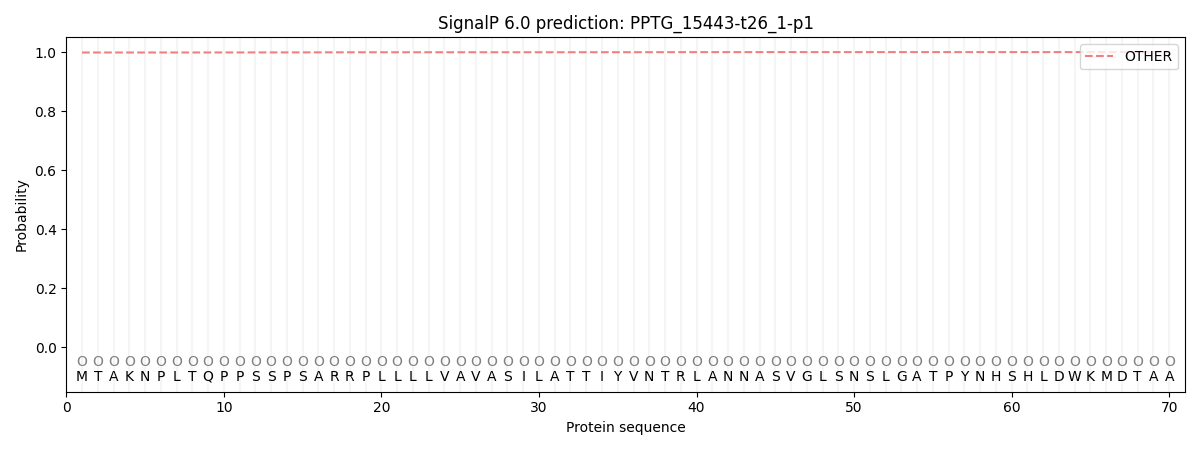
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998751 | 0.001265 |