You are browsing environment: FUNGIDB
CAZyme Information: PPTG_15283-t26_1-p1
You are here: Home > Sequence: PPTG_15283-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_15283-t26_1-p1 | |||||||||||
| CAZy Family | GT1|GT1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.164:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 38 | 504 | 9.2e-108 | 0.9824561403508771 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 405300 | Glyco_hydr_30_2 | 6.99e-14 | 64 | 275 | 39 | 220 | O-Glycosyl hydrolase family 30. |
| 227807 | XynC | 3.38e-08 | 168 | 504 | 117 | 426 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.81e-143 | 12 | 506 | 13 | 496 | |
| 3.64e-111 | 18 | 506 | 29 | 494 | |
| 3.75e-111 | 42 | 506 | 48 | 495 | |
| 3.75e-111 | 42 | 506 | 48 | 495 | |
| 3.72e-110 | 32 | 507 | 32 | 493 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.51e-89 | 3 | 506 | 2 | 473 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
|
| 1.66e-08 | 43 | 266 | 50 | 295 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
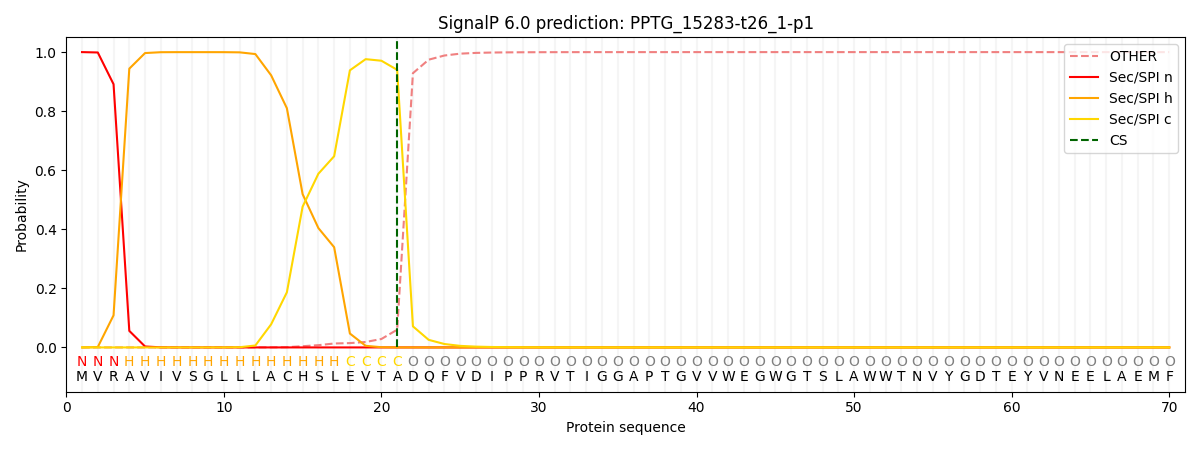
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000389 | 0.999589 | CS pos: 21-22. Pr: 0.9399 |
