You are browsing environment: FUNGIDB
CAZyme Information: PPTG_14561-t26_1-p1
You are here: Home > Sequence: PPTG_14561-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_14561-t26_1-p1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | hypothetical protein, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.256:16 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT59 | 42 | 408 | 1.6e-108 | 0.8910891089108911 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398537 | DIE2_ALG10 | 5.11e-116 | 43 | 409 | 1 | 345 | DIE2/ALG10 family. The ALG10 protein from Saccharomyces cerevisiae encodes the alpha-1,2 glucosyltransferase of the endoplasmic reticulum. This protein has been characterized in rat as potassium channel regulator 1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.91e-95 | 32 | 408 | 14 | 379 | |
| 5.53e-74 | 38 | 407 | 26 | 389 | |
| 5.53e-74 | 38 | 407 | 26 | 389 | |
| 5.53e-74 | 38 | 407 | 26 | 389 | |
| 2.06e-73 | 35 | 407 | 23 | 387 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.82e-75 | 38 | 407 | 26 | 389 | Putative Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Rattus norvegicus OX=10116 GN=Alg10b PE=1 SV=1 |
|
| 2.98e-73 | 38 | 407 | 26 | 389 | Putative Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Mus musculus OX=10090 GN=Alg10b PE=1 SV=1 |
|
| 1.24e-71 | 38 | 407 | 26 | 388 | Putative Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Homo sapiens OX=9606 GN=ALG10B PE=1 SV=2 |
|
| 1.74e-71 | 38 | 407 | 26 | 388 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Homo sapiens OX=9606 GN=ALG10 PE=1 SV=1 |
|
| 1.56e-51 | 25 | 398 | 4 | 412 | Dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase OS=Arabidopsis thaliana OX=3702 GN=ALG10 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000020 | 0.000007 |
TMHMM Annotations download full data without filtering help
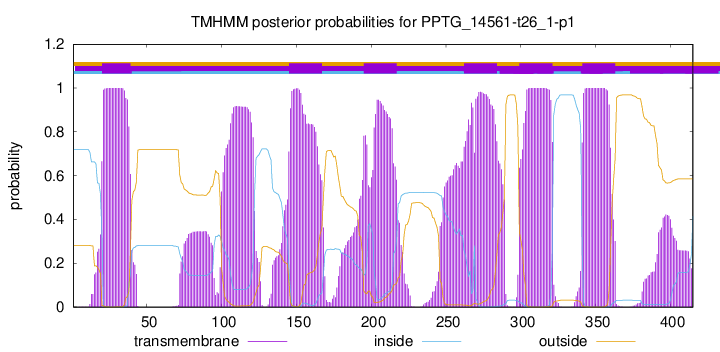
| Start | End |
|---|---|
| 20 | 39 |
| 145 | 167 |
| 195 | 217 |
| 262 | 284 |
| 299 | 321 |
| 341 | 363 |
