You are browsing environment: FUNGIDB
CAZyme Information: PPTG_14134-t26_1-p1
You are here: Home > Sequence: PPTG_14134-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_14134-t26_1-p1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.122:14 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 250845 | Galactosyl_T | 3.26e-22 | 201 | 387 | 1 | 193 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
| 215596 | PLN03133 | 1.04e-11 | 178 | 379 | 377 | 579 | beta-1,3-galactosyltransferase; Provisional |
| 238121 | WD40 | 1.86e-10 | 469 | 808 | 6 | 243 | WD40 domain, found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly; typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40; between GH and WD lies a conserved core; serves as a stable propeller-like platform to which proteins can bind either stably or reversibly; forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel b-sheet; instances with few detectable copies are hypothesized to form larger structures by dimerization; each WD40 sequence repeat forms the first three strands of one blade and the last strand in the next blade; the last C-terminal WD40 repeat completes the blade structure of the first WD40 repeat to create the closed ring propeller-structure; residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands; 7 copies of the repeat are present in this alignment. |
| 225201 | WD40 | 3.08e-08 | 503 | 925 | 39 | 430 | WD40 repeat [General function prediction only]. |
| 238121 | WD40 | 1.79e-07 | 456 | 695 | 73 | 278 | WD40 domain, found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly; typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40; between GH and WD lies a conserved core; serves as a stable propeller-like platform to which proteins can bind either stably or reversibly; forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel b-sheet; instances with few detectable copies are hypothesized to form larger structures by dimerization; each WD40 sequence repeat forms the first three strands of one blade and the last strand in the next blade; the last C-terminal WD40 repeat completes the blade structure of the first WD40 repeat to create the closed ring propeller-structure; residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands; 7 copies of the repeat are present in this alignment. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.45e-17 | 182 | 418 | 431 | 665 | |
| 2.45e-17 | 182 | 418 | 431 | 665 | |
| 2.47e-17 | 188 | 428 | 82 | 323 | |
| 3.25e-17 | 182 | 418 | 437 | 671 | |
| 3.88e-17 | 163 | 394 | 50 | 277 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.88e-16 | 188 | 428 | 80 | 321 | Beta-1,3-galactosyltransferase 1 OS=Mus musculus OX=10090 GN=B3galt1 PE=1 SV=2 |
|
| 4.88e-16 | 188 | 428 | 80 | 321 | Beta-1,3-galactosyltransferase 1 OS=Homo sapiens OX=9606 GN=B3GALT1 PE=1 SV=1 |
|
| 4.88e-16 | 188 | 428 | 80 | 321 | Beta-1,3-galactosyltransferase 1 OS=Pongo pygmaeus OX=9600 GN=B3GALT1 PE=3 SV=1 |
|
| 4.88e-16 | 188 | 428 | 80 | 321 | Beta-1,3-galactosyltransferase 1 OS=Pan troglodytes OX=9598 GN=B3GALT1 PE=3 SV=1 |
|
| 4.88e-16 | 188 | 428 | 80 | 321 | Beta-1,3-galactosyltransferase 1 OS=Pan paniscus OX=9597 GN=B3GALT1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
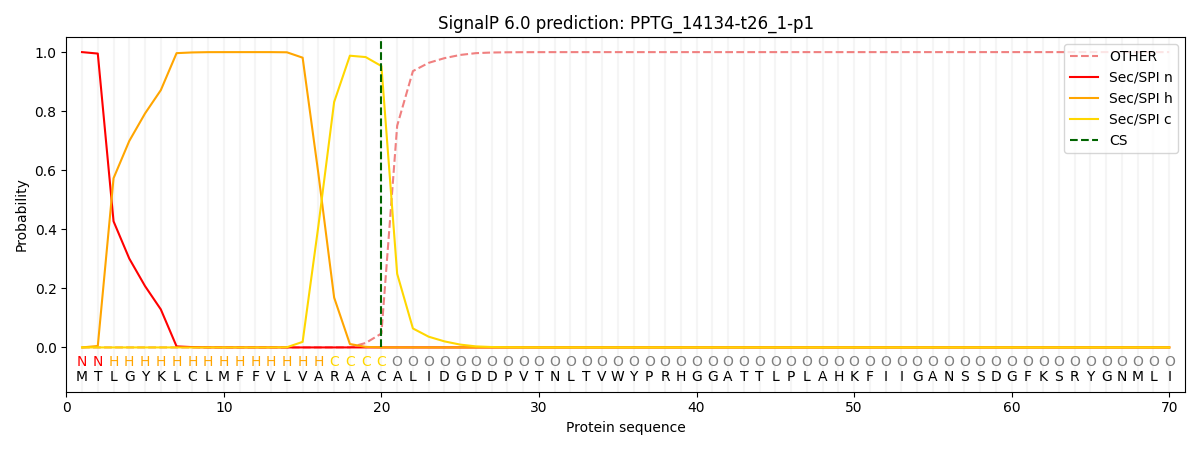
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000236 | 0.999725 | CS pos: 20-21. Pr: 0.9527 |

