You are browsing environment: FUNGIDB
CAZyme Information: PPTG_14132-t26_1-p1
You are here: Home > Sequence: PPTG_14132-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_14132-t26_1-p1 | |||||||||||
| CAZy Family | GH81 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT10 | 731 | 890 | 4.4e-34 | 0.42939481268011526 |
| GT31 | 223 | 411 | 2.3e-29 | 0.9791666666666666 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395683 | Glyco_transf_10 | 4.34e-15 | 761 | 885 | 17 | 130 | Glycosyltransferase family 10 (fucosyltransferase) C-term. This is the C-terminal domain of a family of fucosyltransferases. This enzyme transfers fucose from GDP-Fucose to GlcNAc in an alpha1,3 linkage. This family is known as glycosyltransferase family 10. The C-terminal domain is the likely binding-region for ADP (manuscript in publication). |
| 250845 | Galactosyl_T | 3.15e-13 | 223 | 411 | 3 | 194 | Galactosyltransferase. This family includes the galactosyltransferases UDP-galactose:2-acetamido-2-deoxy-D-glucose3beta-galactosyltransferase and UDP-Gal:beta-GlcNAc beta 1,3-galactosyltranferase. Specific galactosyltransferases transfer galactose to GlcNAc terminal chains in the synthesis of the lacto-series oligosaccharides types 1 and 2. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.03e-19 | 626 | 892 | 23 | 267 | |
| 5.10e-19 | 745 | 892 | 124 | 265 | |
| 5.10e-19 | 745 | 892 | 124 | 265 | |
| 5.10e-19 | 745 | 892 | 124 | 265 | |
| 6.41e-19 | 713 | 892 | 43 | 211 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.07e-13 | 743 | 897 | 176 | 316 | Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_B Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_C Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZX_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZY_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori] |
|
| 1.57e-12 | 743 | 897 | 176 | 316 | Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],5ZOI_B Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.26e-18 | 713 | 892 | 398 | 566 | Putative fucosyltransferase R654 OS=Acanthamoeba polyphaga mimivirus OX=212035 GN=MIMI_R654 PE=3 SV=1 |
|
| 1.91e-12 | 743 | 897 | 176 | 316 | Alpha-(1,3)-fucosyltransferase FucT OS=Helicobacter pylori OX=210 GN=fucT PE=1 SV=1 |
|
| 1.25e-10 | 755 | 887 | 222 | 359 | Alpha-(1,4)-fucosyltransferase OS=Arabidopsis thaliana OX=3702 GN=FUT13 PE=2 SV=2 |
|
| 2.84e-10 | 587 | 887 | 68 | 339 | Alpha-(1,3)-fucosyltransferase 10 OS=Xenopus laevis OX=8355 GN=fut10 PE=2 SV=2 |
|
| 8.04e-10 | 752 | 896 | 175 | 324 | Alpha-(1,3)-fucosyltransferase B OS=Drosophila melanogaster OX=7227 GN=FucTB PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
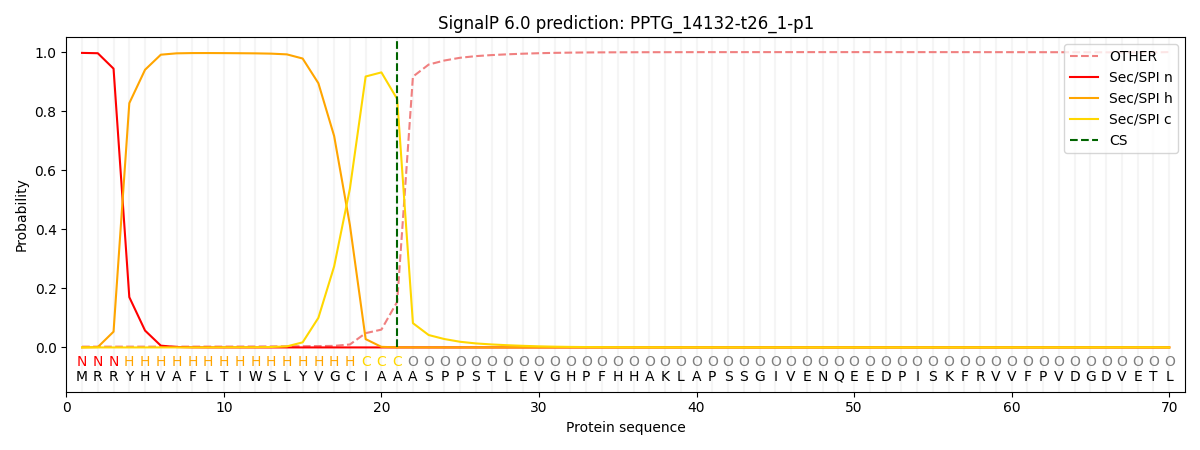
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.005533 | 0.994444 | CS pos: 21-22. Pr: 0.8412 |
