You are browsing environment: FUNGIDB
CAZyme Information: PPTG_12006-t26_1-p1
You are here: Home > Sequence: PPTG_12006-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
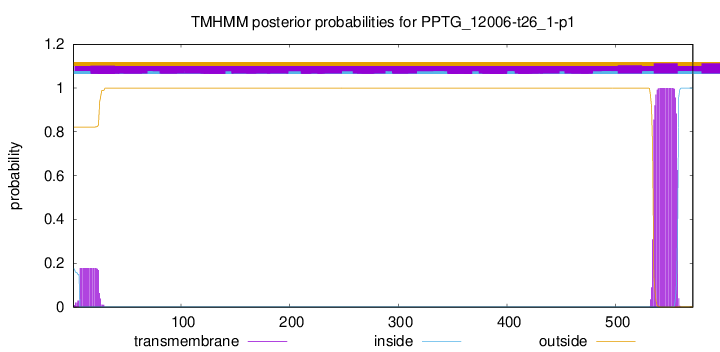
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_12006-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:2 | 3.2.1.23:1 | 3.2.1.38:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 28 | 509 | 8.3e-136 | 0.9836829836829837 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 7.01e-145 | 29 | 510 | 5 | 451 | Glycosyl hydrolase family 1. |
| 225343 | BglB | 4.21e-117 | 29 | 513 | 4 | 456 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215435 | PLN02814 | 1.05e-92 | 7 | 520 | 6 | 488 | beta-glucosidase |
| 215455 | PLN02849 | 8.69e-90 | 10 | 503 | 12 | 476 | beta-glucosidase |
| 184102 | PRK13511 | 1.95e-77 | 29 | 510 | 5 | 466 | 6-phospho-beta-galactosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 6.61e-270 | 11 | 557 | 14 | 561 | |
| 1.00e-265 | 22 | 557 | 6 | 537 | |
| 1.21e-208 | 23 | 513 | 17 | 507 | |
| 1.85e-151 | 77 | 515 | 5 | 421 | |
| 3.46e-106 | 29 | 510 | 36 | 511 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.82e-101 | 8 | 505 | 13 | 503 | Crystal structure of beta-primeverosidase [Camellia sinensis],3WQ4_B Crystal structure of beta-primeverosidase [Camellia sinensis],3WQ5_A beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, natural aglycone derivative [Camellia sinensis],3WQ5_B beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, natural aglycone derivative [Camellia sinensis],3WQ6_A beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, artificial aglycone derivative [Camellia sinensis],3WQ6_B beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, artificial aglycone derivative [Camellia sinensis] |
|
| 7.68e-97 | 22 | 505 | 23 | 501 | Chain A, Beta-glucosidase Os4BGlu12 [Oryza sativa],3PTK_B Chain B, Beta-glucosidase Os4BGlu12 [Oryza sativa],3PTM_A Chain A, Beta-glucosidase Os4BGlu12 [Oryza sativa],3PTM_B Chain B, Beta-glucosidase Os4BGlu12 [Oryza sativa],3PTQ_A Chain A, Beta-glucosidase Os4BGlu12 [Oryza sativa],3PTQ_B Chain B, Beta-glucosidase Os4BGlu12 [Oryza sativa] |
|
| 2.21e-95 | 27 | 510 | 36 | 500 | Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase [Oryza sativa Indica Group],4JIE_A Structural analysis and insights into glycon specificity of the rice GH1 Os7BGlu26 beta-D-mannosidase [Oryza sativa Indica Group] |
|
| 2.21e-95 | 27 | 510 | 36 | 500 | Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase [Oryza sativa Indica Group],4RE3_A Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase [Oryza sativa Indica Group],4RE4_A Different transition state conformations for the hydrolysis of beta-mannosides and beta-glucosides in the rice Os7BGlu26 family GH1 beta-mannosidase/beta-glucosidase [Oryza sativa Indica Group] |
|
| 2.48e-93 | 13 | 502 | 6 | 506 | Crystal structure of native Raucaffricine glucosidase [Rauvolfia serpentina],4ATD_B Crystal structure of native Raucaffricine glucosidase [Rauvolfia serpentina],4ATL_A Crystal structure of Raucaffricine glucosidase in complex with Glucose [Rauvolfia serpentina],4ATL_B Crystal structure of Raucaffricine glucosidase in complex with Glucose [Rauvolfia serpentina] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.09e-104 | 29 | 504 | 37 | 506 | Beta-glucosidase 32 OS=Arabidopsis thaliana OX=3702 GN=BGLU32 PE=2 SV=2 |
|
| 1.44e-103 | 29 | 532 | 31 | 526 | Beta-glucosidase 30 OS=Arabidopsis thaliana OX=3702 GN=BGLU30 PE=2 SV=1 |
|
| 1.28e-102 | 29 | 546 | 34 | 542 | Beta-glucosidase 28 OS=Arabidopsis thaliana OX=3702 GN=BGLU28 PE=2 SV=1 |
|
| 1.03e-101 | 29 | 508 | 37 | 510 | Beta-glucosidase 31 OS=Arabidopsis thaliana OX=3702 GN=BGLU31 PE=2 SV=1 |
|
| 3.72e-100 | 29 | 505 | 19 | 487 | Beta-glucosidase 27 OS=Arabidopsis thaliana OX=3702 GN=BGLU27 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
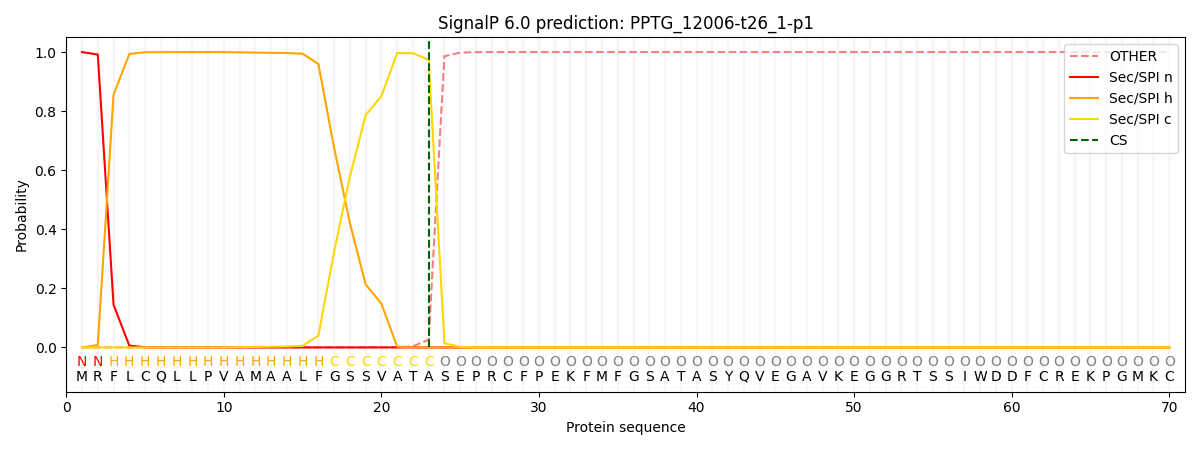
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000232 | 0.999762 | CS pos: 23-24. Pr: 0.9737 |