You are browsing environment: FUNGIDB
CAZyme Information: PPTG_11791-t26_1-p1
You are here: Home > Sequence: PPTG_11791-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_11791-t26_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | hypothetical protein, variant 3 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM47 | 1122 | 1261 | 1.2e-18 | 0.9609375 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227511 | ATS1 | 1.87e-22 | 837 | 1104 | 187 | 440 | Alpha-tubulin suppressor and related RCC1 domain-containing proteins [Cell cycle control, cell division, chromosome partitioning, Cytoskeleton]. |
| 227511 | ATS1 | 2.55e-21 | 847 | 1090 | 125 | 374 | Alpha-tubulin suppressor and related RCC1 domain-containing proteins [Cell cycle control, cell division, chromosome partitioning, Cytoskeleton]. |
| 227455 | FRQ1 | 1.17e-19 | 81 | 233 | 7 | 156 | Ca2+-binding protein, EF-hand superfamily [Signal transduction mechanisms]. |
| 227511 | ATS1 | 2.37e-16 | 846 | 1095 | 69 | 323 | Alpha-tubulin suppressor and related RCC1 domain-containing proteins [Cell cycle control, cell division, chromosome partitioning, Cytoskeleton]. |
| 238008 | EFh | 3.88e-16 | 172 | 231 | 3 | 62 | EF-hand, calcium binding motif; A diverse superfamily of calcium sensors and calcium signal modulators; most examples in this alignment model have 2 active canonical EF hands. Ca2+ binding induces a conformational change in the EF-hand motif, leading to the activation or inactivation of target proteins. EF-hands tend to occur in pairs or higher copy numbers. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.23e-13 | 1112 | 1266 | 203 | 340 | |
| 4.23e-13 | 1112 | 1266 | 203 | 340 | |
| 2.13e-12 | 1109 | 1279 | 144 | 305 | |
| 2.99e-12 | 1112 | 1266 | 131 | 272 | |
| 6.88e-12 | 1018 | 1266 | 30 | 272 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.49e-25 | 832 | 1109 | 112 | 372 | Chain A, Ultraviolet-B receptor UVR8 [Arabidopsis thaliana],6XZN_B Chain B, Ultraviolet-B receptor UVR8 [Arabidopsis thaliana] |
|
| 6.49e-25 | 804 | 1089 | 33 | 301 | Chain A, Ultraviolet-B receptor UVR8 [Arabidopsis thaliana],6XZM_B Chain B, Ultraviolet-B receptor UVR8 [Arabidopsis thaliana] |
|
| 1.39e-24 | 824 | 1123 | 118 | 393 | Crystal structure of the third RCC1-like domain of HERC1 [Homo sapiens],4O2W_B Crystal structure of the third RCC1-like domain of HERC1 [Homo sapiens],4O2W_C Crystal structure of the third RCC1-like domain of HERC1 [Homo sapiens],4O2W_D Crystal structure of the third RCC1-like domain of HERC1 [Homo sapiens] |
|
| 1.51e-24 | 832 | 1109 | 109 | 369 | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 [Arabidopsis thaliana],4DNV_B Crystal structure of the W285F mutant of UVB-resistance protein UVR8 [Arabidopsis thaliana],4DNV_C Crystal structure of the W285F mutant of UVB-resistance protein UVR8 [Arabidopsis thaliana],4DNV_D Crystal structure of the W285F mutant of UVB-resistance protein UVR8 [Arabidopsis thaliana] |
|
| 1.61e-24 | 804 | 1089 | 30 | 298 | Crystal structure of UVB-resistance protein UVR8 [Arabidopsis thaliana],4DNW_B Crystal structure of UVB-resistance protein UVR8 [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.70e-26 | 847 | 1086 | 43 | 265 | E3 ISG15--protein ligase Herc6 OS=Mus musculus OX=10090 GN=Herc6 PE=2 SV=1 |
|
| 4.59e-25 | 841 | 1093 | 38 | 273 | Probable E3 ubiquitin-protein ligase HERC6 OS=Homo sapiens OX=9606 GN=HERC6 PE=1 SV=2 |
|
| 2.06e-23 | 804 | 1089 | 41 | 309 | Ultraviolet-B receptor UVR8 OS=Arabidopsis thaliana OX=3702 GN=UVR8 PE=1 SV=1 |
|
| 1.82e-22 | 824 | 1123 | 4074 | 4349 | Probable E3 ubiquitin-protein ligase HERC1 OS=Homo sapiens OX=9606 GN=HERC1 PE=1 SV=2 |
|
| 2.38e-22 | 805 | 1182 | 2966 | 3351 | E3 ubiquitin-protein ligase HERC2 OS=Mus musculus OX=10090 GN=Herc2 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
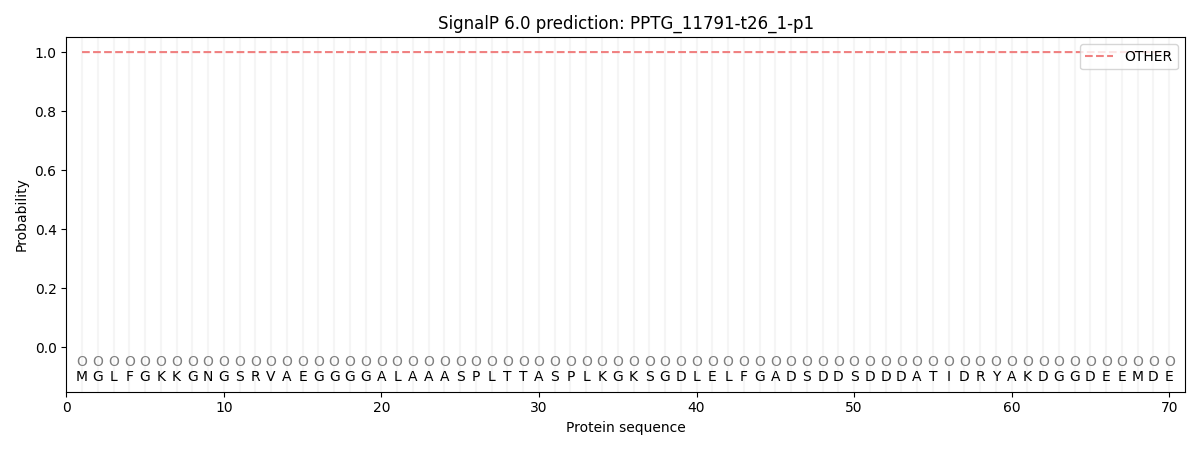
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000004 |
