You are browsing environment: FUNGIDB
CAZyme Information: PPTG_11180-t26_2-p1
You are here: Home > Sequence: PPTG_11180-t26_2-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_11180-t26_2-p1 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | hypothetical protein, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.99.18:21 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT66 | 13 | 598 | 1e-127 | 0.7417027417027418 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396873 | STT3 | 2.03e-71 | 17 | 332 | 90 | 410 | Oligosaccharyl transferase STT3 subunit. This family consists of the oligosaccharyl transferase STT3 subunit and related proteins. The STT3 subunit is part of the oligosaccharyl transferase (OTase) complex of proteins and is required for its activity. In eukaryotes, OTase transfers a lipid-linked core-oligosaccharide to selected asparagine residues in the ER. In the archaea STT3 occurs alone, rather than in an OTase complex, and is required for N-glycosylation of asparagines. |
| 224206 | Stt3 | 1.65e-49 | 13 | 647 | 102 | 730 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
| 275016 | archaeo_STT3 | 1.16e-18 | 20 | 527 | 93 | 584 | oligosaccharyl transferase, archaeosortase A system-associated. Members of this protein family occur, one to three members per genome, in the same species of Euryarchaeota as contain the predicted protein-sorting enzyme archaeosortase (TIGR04125) and its cognate protein-sorting signal PGF-CTERM (TIGR04126). |
| 404170 | PMT_2 | 1.06e-04 | 9 | 149 | 8 | 146 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.87e-196 | 1 | 646 | 79 | 741 | |
| 1.87e-196 | 1 | 646 | 79 | 741 | |
| 1.47e-189 | 4 | 648 | 139 | 737 | |
| 1.47e-189 | 4 | 648 | 139 | 737 | |
| 2.64e-188 | 4 | 648 | 140 | 738 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.16e-159 | 1 | 647 | 147 | 773 | Cryo-EM structure of human oligosaccharyltransferase complex OST-B [Homo sapiens] |
|
| 1.41e-154 | 16 | 647 | 107 | 695 | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome [Canis lupus familiaris] |
|
| 1.84e-154 | 16 | 647 | 107 | 695 | Subtomogram average of OST-containing ribosome-translocon complexes from canine rough microsomal membranes [Canis lupus familiaris],6FTI_5 Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Programmed 80S Ribosome [Canis lupus familiaris] |
|
| 1.84e-154 | 16 | 647 | 107 | 695 | Cryo-EM structure of human oligosaccharyltransferase complex OST-A [Homo sapiens] |
|
| 9.94e-135 | 15 | 648 | 105 | 684 | The Cryo-EM structure of a eukaryotic oligosaccharyl transferase complex [Saccharomyces cerevisiae S288C],6EZN_F Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex [Saccharomyces cerevisiae S288C],7OCI_F Chain F, Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3 [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.23e-159 | 1 | 647 | 147 | 773 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS=Canis lupus familiaris OX=9615 GN=STT3B PE=1 SV=1 |
|
| 5.97e-159 | 1 | 647 | 147 | 773 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS=Homo sapiens OX=9606 GN=STT3B PE=1 SV=1 |
|
| 1.33e-158 | 1 | 647 | 116 | 729 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS=Arabidopsis thaliana OX=3702 GN=STT3B PE=2 SV=1 |
|
| 2.17e-158 | 1 | 647 | 144 | 770 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B OS=Mus musculus OX=10090 GN=Stt3b PE=1 SV=2 |
|
| 4.01e-157 | 15 | 648 | 108 | 717 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit stt-3 OS=Caenorhabditis elegans OX=6239 GN=stt-3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
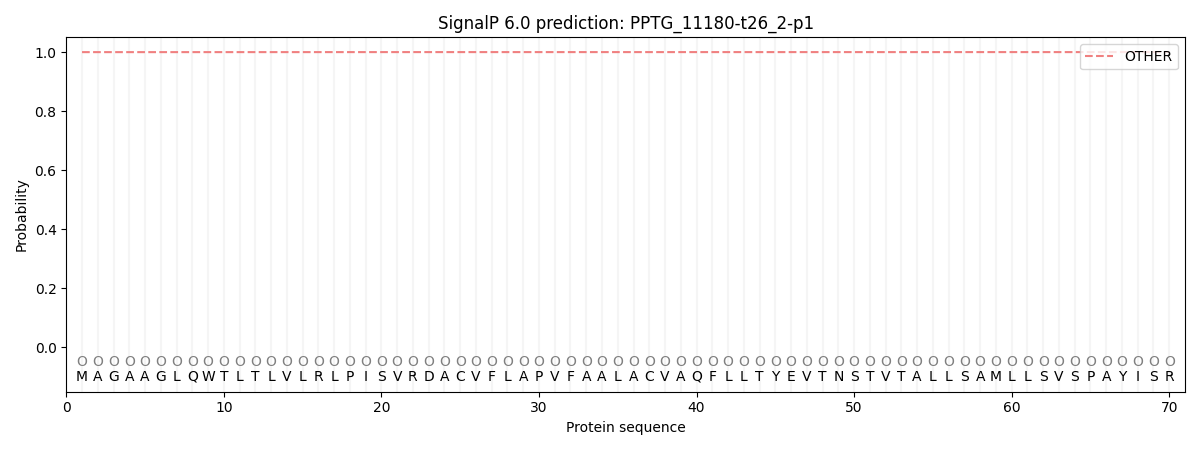
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999975 | 0.000039 |
TMHMM Annotations download full data without filtering help
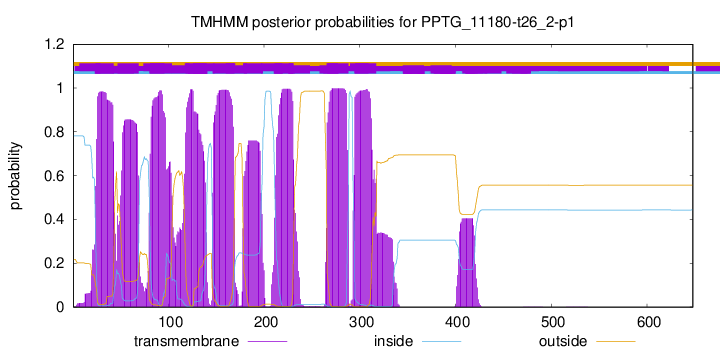
| Start | End |
|---|---|
| 24 | 43 |
| 47 | 69 |
| 81 | 103 |
| 118 | 140 |
| 147 | 169 |
| 179 | 198 |
| 211 | 230 |
| 266 | 288 |
| 295 | 317 |
