You are browsing environment: FUNGIDB
CAZyme Information: PPTG_09841-t26_1-p1
You are here: Home > Sequence: PPTG_09841-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_09841-t26_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | CMGC/CDK protein kinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH72 | 25 | 289 | 9.2e-66 | 0.7852564102564102 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397351 | Glyco_hydro_72 | 1.79e-54 | 17 | 288 | 1 | 251 | Glucanosyltransferase. This is a family of glycosylphosphatidylinositol-anchored beta(1-3)glucanosyltransferases. The active site residues in the Aspergillus fumigatus example are the two glutamate residues at 160 and 261. |
| 380950 | SET_NSD | 6.60e-54 | 415 | 555 | 1 | 142 | SET domain (including post-SET domain) found in nuclear SET domain-containing proteins, NSD1, NSD2, NSD3 and similar proteins. The nuclear receptor-binding SET Domain (NSD) family of histone H3 lysine 36 methyltransferases is comprised of NSD1, NSD2, and NSD3, which are primarily known to be involved in chromatin integrity and gene expression through mono-, di-, or tri-methylating lysine 36 of histone H3 (H3K36), respectively. NSD1 (EC 2.1.1.43; also termed histone-lysine N-methyltransferase H3 lysine-36 and H4 lysine-20 specific, androgen receptor coactivator 267 kDa protein (ARA267), androgen receptor-associated protein of 267 kDa, H3-K36-HMTase, H4-K20-HMTase, lysine N-methyltransferase 3B (KMT3B) or NR-binding SET domain-containing protein 1) functions as a histone-lysine N-methyltransferase that preferentially methylates 'Lys-36' of histone H3 and 'Lys-20' of histone H4. NSD2 (EC 2.1.1.43; also termed multiple myeloma SET domain-containing protein (MMSET), protein trithorax-5 (TRX5), or wolf-Hirschhorn syndrome candidate 1 protein (WHSC1)) acts as histone-lysine N-methyltransferase with histone H3 'Lys-27' (H3K27me) methyltransferase activity. NSD3 (EC 2.1.1.43; also termed protein whistle, WHSC1-like 1 isoform 9 with methyltransferase activity to lysine, Wolf-Hirschhorn syndrome candidate 1-like protein 1 (WHSC1L1), or WHSC1-like protein 1) functions as a histone-lysine N-methyltransferase that preferentially methylates 'Lys-4' and 'Lys-27' of histone H3. |
| 380951 | SET_ASH1L | 5.11e-53 | 417 | 556 | 1 | 141 | SET domain (including post-SET domain) found in ASH1-like protein (ASH1L) and similar proteins. ASH1L (EC 2.1.1.43; also termed absent small and homeotic disks protein 1 homolog, KMT2H, or lysine N-methyltransferase 2H) acts as histone-lysine N-methyltransferase that specifically methylates 'Lys-36' of histone H3 (H3K36me). It plays important roles in development; heterozygous mutation of ASH1L is associated with severe intellectual disability (ID) and multiple congenital anomaly (MCA). |
| 380929 | SET_SETD2-like | 9.20e-53 | 418 | 551 | 2 | 136 | SET domain (including post-SET domain) found in SET domain-containing protein 2 (SETD2), nuclear SETD2 (NSD2), ASH1-like protein (ASH1L) and similar proteins. This family includes SET domain-containing protein 2 (SETD2), nuclear SETD2 (NSD2) and ASH1-like protein (ASH1L), which function as histone-lysine N-methyltransferases. SETD2 specifically trimethylates 'Lys-36' of histone H3 (H3K36me3) using demethylated 'Lys-36' (H3K36me2) as substrate. NSD2 shows histone H3 'Lys-27' (H3K27me) methyltransferase activity. ASH1L specifically methylates 'Lys-36' of histone H3 (H3K36me). The family also includes Arabidopsis thaliana ASH1-related protein 3 (ASHR3) and similar proteins. |
| 380949 | SET_SETD2 | 4.99e-52 | 415 | 555 | 1 | 142 | SET domain (including post-SET domain) found in SET domain-containing protein 2 (SETD2) and similar proteins. SETD2 (also termed HIF-1, huntingtin yeast partner B, huntingtin-interacting protein 1 (HIP-1), huntingtin-interacting protein B, lysine N-methyltransferase 3A or protein-lysine N-methyltransferase SETD2) acts as histone-lysine N-methyltransferase that specifically trimethylates 'Lys-36' of histone H3 (H3K36me3) using demethylated 'Lys-36' (H3K36me2) as substrate. It has been shown that methylation is a posttranslational modification of dynamic microtubules and that SETD2 methylates alpha-tubulin at lysine 40, the same lysine that is marked by acetylation on microtubules. Methylation of microtubules occurs during mitosis and cytokinesis and can be ablated by SETD2 deletion, which causes mitotic spindle and cytokinesis defects, micronuclei, and polyploidy. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 8 | 1788 | 505 | 2516 | |
| 5.91e-103 | 19 | 312 | 37 | 335 | |
| 2.43e-101 | 22 | 300 | 24 | 302 | |
| 7.21e-97 | 24 | 300 | 25 | 301 | |
| 9.68e-93 | 23 | 305 | 2186 | 2463 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.77e-46 | 323 | 558 | 29 | 247 | crystal structure of SESTD2 in complex with H3.3S31phK36M peptide [Homo sapiens] |
|
| 6.67e-46 | 323 | 558 | 24 | 242 | Chain L, Histone-lysine N-methyltransferase SETD2 [Homo sapiens] |
|
| 1.74e-45 | 323 | 558 | 42 | 260 | Crystal structure of Methyltransferase domain of human SET domain-containing protein 2 Compound: Pr-SNF [Homo sapiens],4H12_A The crystal structure of methyltransferase domain of human SET domain-containing protein 2 in complex with S-adenosyl-L-homocysteine [Homo sapiens],7LZB_A Chain A, Histone-lysine N-methyltransferase SETD2 [Homo sapiens],7LZD_A Chain A, Histone-lysine N-methyltransferase SETD2 [Homo sapiens],7LZF_A Chain A, Histone-lysine N-methyltransferase SETD2 [Homo sapiens] |
|
| 1.79e-45 | 323 | 558 | 43 | 261 | Crystal structure of SETD2 bound to histone H3.3 K36M peptide [Homo sapiens],5JLB_A Crystal structure of SETD2 bound to histone H3.3 K36I peptide [Homo sapiens],5JLE_A Crystal structure of SETD2 bound to SAH [Homo sapiens] |
|
| 2.80e-45 | 323 | 558 | 59 | 277 | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],5LSX_A Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],5LSY_A Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],5LSZ_A Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],5LT6_A Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],5LT6_B Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],5LT7_A Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],5LT8_A Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives [Homo sapiens],6VDB_A SETD2 in complex with a H3-variant peptide [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.37e-40 | 323 | 583 | 1449 | 1693 | Histone-lysine N-methyltransferase SETD2 OS=Mus musculus OX=10090 GN=Setd2 PE=1 SV=1 |
|
| 3.12e-40 | 323 | 583 | 1475 | 1719 | Histone-lysine N-methyltransferase SETD2 OS=Homo sapiens OX=9606 GN=SETD2 PE=1 SV=3 |
|
| 3.55e-39 | 382 | 574 | 1806 | 2001 | Histone-lysine N-methyltransferase, H3 lysine-36 specific OS=Mus musculus OX=10090 GN=Nsd1 PE=1 SV=1 |
|
| 9.77e-39 | 373 | 589 | 1347 | 1557 | Histone-lysine N-methyltransferase ash1 OS=Drosophila melanogaster OX=7227 GN=ash1 PE=1 SV=3 |
|
| 1.39e-38 | 382 | 557 | 1908 | 2084 | Histone-lysine N-methyltransferase, H3 lysine-36 specific OS=Homo sapiens OX=9606 GN=NSD1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
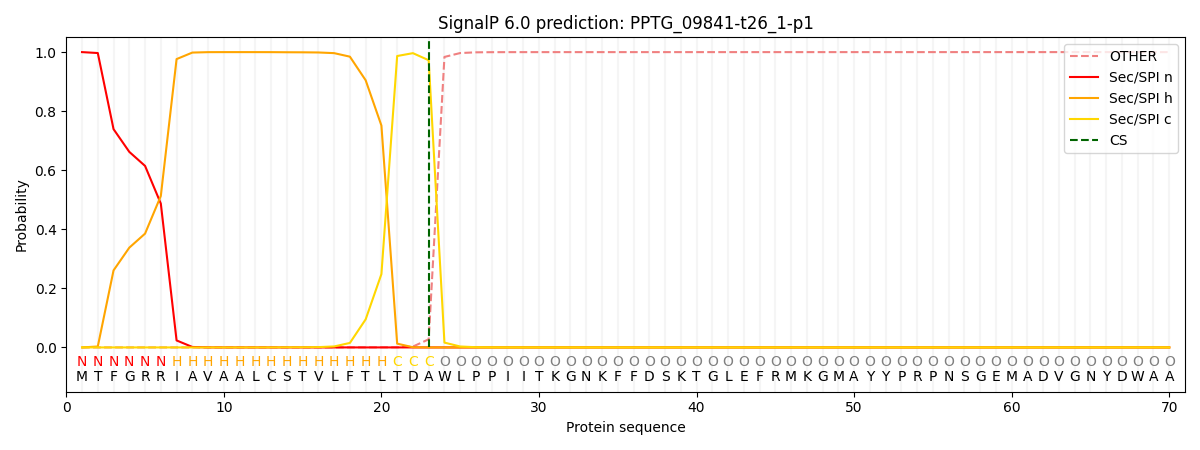
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000636 | 0.999329 | CS pos: 23-24. Pr: 0.9725 |

