You are browsing environment: FUNGIDB
CAZyme Information: PPTG_08947-t26_1-p1
Basic Information
help
| Species |
Phytophthora parasitica
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica
|
| CAZyme ID |
PPTG_08947-t26_1-p1
|
| CAZy Family |
GH2 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 955 |
|
107960.06 |
6.5076 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PparasiticaINRA-310 |
23240 |
761204 |
119 |
23121
|
|
| Gene Location |
No EC number prediction in PPTG_08947-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GT96 |
521 |
651 |
1.2e-20 |
0.36156351791530944 |
PPTG_08947-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 0.0 |
39 |
950 |
1 |
913 |
| 4.09e-26 |
515 |
926 |
111 |
518 |
| 3.67e-15 |
520 |
901 |
1 |
307 |
| 9.85e-14 |
518 |
678 |
32 |
175 |
| 1.32e-13 |
520 |
800 |
37 |
277 |
PPTG_08947-t26_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.28e-13 |
511 |
790 |
28 |
264 |
Peptidyl serine alpha-galactosyltransferase OS=Chlamydomonas reinhardtii OX=3055 GN=SGT1 PE=1 SV=1 |
| 1.76e-11 |
513 |
809 |
23 |
255 |
Peptidyl serine alpha-galactosyltransferase OS=Arabidopsis thaliana OX=3702 GN=SERGT1 PE=2 SV=1 |
This protein is predicted as OTHER
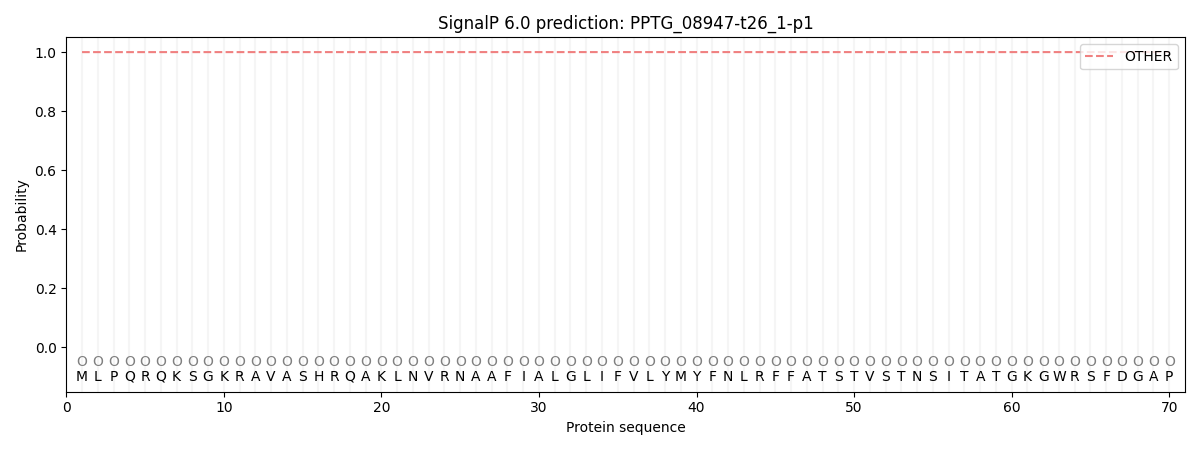
| Other |
SP_Sec_SPI |
CS Position |
| 1.000004 |
0.000011 |
|