You are browsing environment: FUNGIDB
CAZyme Information: PPTG_07182-t26_1-p1
You are here: Home > Sequence: PPTG_07182-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_07182-t26_1-p1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 27 | 193 | 8e-37 | 0.9788359788359788 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 2.68e-30 | 27 | 184 | 1 | 162 | Cutinase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.25e-118 | 1 | 205 | 1 | 217 | |
| 7.11e-90 | 16 | 204 | 14 | 214 | |
| 8.91e-86 | 16 | 198 | 14 | 208 | |
| 7.45e-82 | 1 | 205 | 1 | 223 | |
| 7.45e-82 | 1 | 205 | 1 | 223 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.67e-40 | 26 | 197 | 34 | 219 | Cutinase OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=cut PE=1 SV=1 |
|
| 9.30e-27 | 21 | 202 | 37 | 228 | Probable carboxylesterase Culp3 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut3 PE=1 SV=1 |
|
| 9.30e-27 | 21 | 202 | 37 | 228 | Probable carboxylesterase Culp3 OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=cut3 PE=3 SV=1 |
|
| 1.39e-25 | 28 | 170 | 44 | 198 | Probable carboxylesterase Culp3 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=cut3 PE=3 SV=1 |
|
| 4.94e-25 | 11 | 179 | 28 | 210 | Phospholipase Culp4 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut4 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
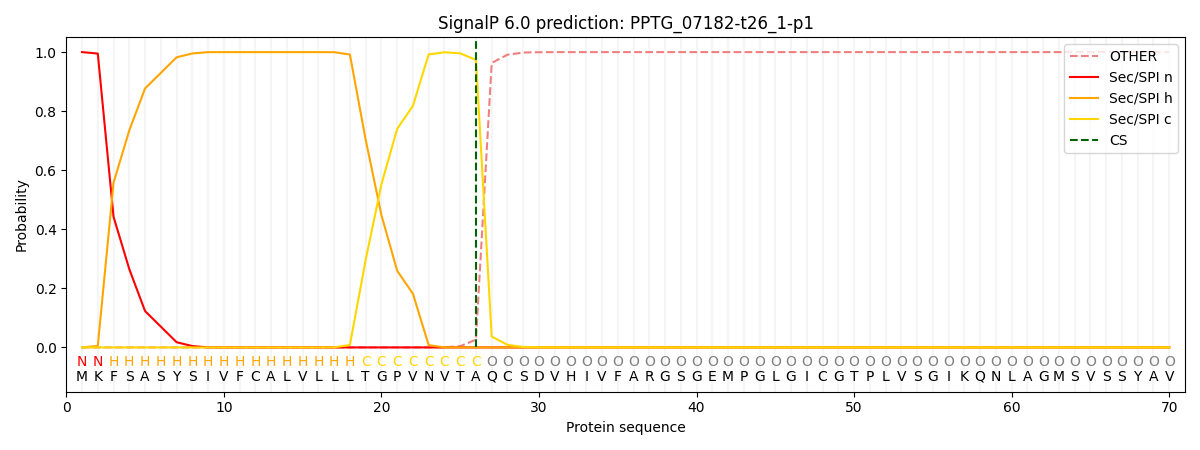
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000193 | 0.999797 | CS pos: 26-27. Pr: 0.9737 |

