You are browsing environment: FUNGIDB
CAZyme Information: PPTG_06233-t26_1-p1
You are here: Home > Sequence: PPTG_06233-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_06233-t26_1-p1 | |||||||||||
| CAZy Family | GH12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.11:1 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 178051 | PLN02432 | 8.45e-22 | 40 | 238 | 13 | 190 | putative pectinesterase |
| 215367 | PLN02682 | 2.04e-18 | 31 | 236 | 63 | 263 | pectinesterase family protein |
| 178372 | PLN02773 | 4.73e-18 | 50 | 241 | 15 | 200 | pectinesterase |
| 178504 | PLN02916 | 7.34e-17 | 34 | 241 | 183 | 380 | pectinesterase family protein |
| 215357 | PLN02665 | 1.62e-16 | 50 | 241 | 78 | 260 | pectinesterase family protein |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.86e-95 | 1 | 241 | 1 | 237 | |
| 9.86e-95 | 1 | 241 | 1 | 237 | |
| 1.95e-92 | 13 | 241 | 10 | 238 | |
| 7.95e-91 | 1 | 239 | 1 | 234 | |
| 2.64e-90 | 1 | 241 | 1 | 241 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.72e-32 | 31 | 239 | 2 | 194 | Crystal Structure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
|
| 2.41e-32 | 31 | 239 | 2 | 194 | Crystal Structure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
|
| 3.52e-11 | 50 | 234 | 13 | 186 | Chain A, Pectinesterase 1 [Solanum lycopersicum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.30e-45 | 31 | 250 | 19 | 232 | Pectinesterase OS=Aspergillus niger OX=5061 GN=pme1 PE=1 SV=1 |
|
| 1.01e-44 | 31 | 250 | 19 | 233 | Pectinesterase OS=Aspergillus aculeatus OX=5053 GN=pme1 PE=2 SV=1 |
|
| 4.34e-36 | 1 | 234 | 1 | 214 | Probable pectinesterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pmeA PE=3 SV=1 |
|
| 1.29e-34 | 31 | 241 | 28 | 222 | Pectinesterase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pmeA PE=1 SV=1 |
|
| 2.62e-33 | 49 | 239 | 41 | 219 | Probable pectinesterase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=pmeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
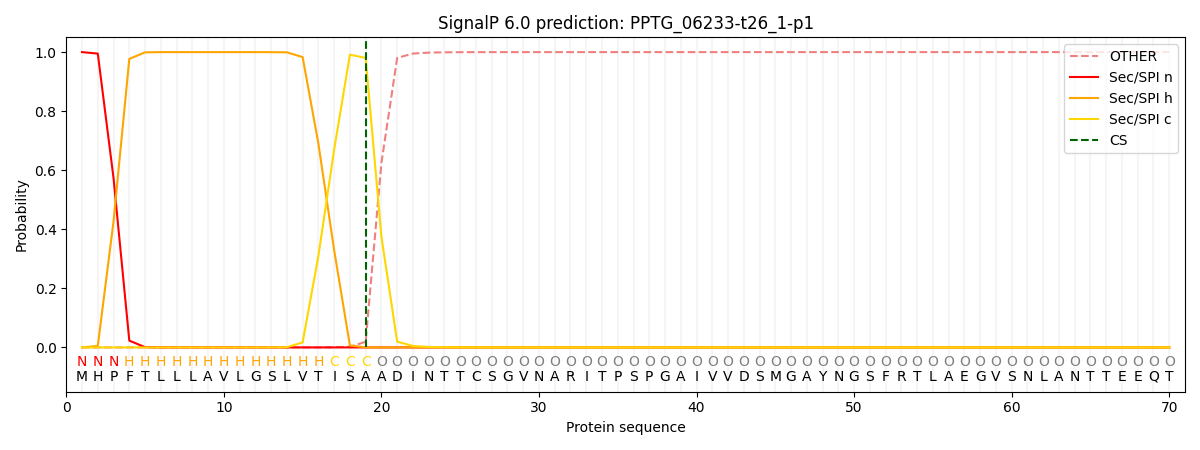
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000202 | 0.999766 | CS pos: 19-20. Pr: 0.9805 |
