You are browsing environment: FUNGIDB
CAZyme Information: PPTG_04305-t26_1-p1
You are here: Home > Sequence: PPTG_04305-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
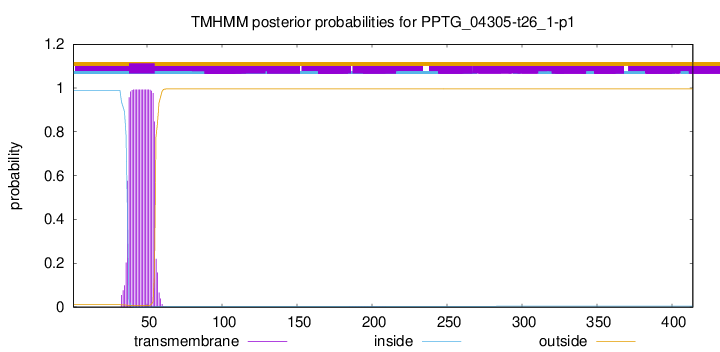
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_04305-t26_1-p1 | |||||||||||
| CAZy Family | CBM9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT62 | 130 | 370 | 1.4e-38 | 0.9664179104477612 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397491 | Anp1 | 2.84e-40 | 129 | 369 | 3 | 262 | Anp1. The members of this family (Anp1, Van1 and Mnn9) are membrane proteins required for proper Golgi function. These proteins co-localize within the cis Golgi, and that they are physically associated in two distinct complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.32e-53 | 135 | 412 | 72 | 375 | |
| 8.32e-53 | 135 | 412 | 72 | 375 | |
| 1.27e-49 | 135 | 412 | 82 | 396 | |
| 1.16e-48 | 145 | 413 | 98 | 392 | |
| 1.16e-48 | 145 | 413 | 98 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.03e-15 | 179 | 413 | 48 | 300 | Crystal structure of Saccharomyces cerevisiae Mnn9 in complex with GDP and Mn. [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.14e-25 | 128 | 412 | 44 | 334 | Mannan polymerase complex subunit mnn9 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mnn9 PE=3 SV=1 |
|
| 8.20e-16 | 213 | 413 | 212 | 419 | Vanadate resistance protein OS=Candida albicans OX=5476 GN=VAN1 PE=3 SV=1 |
|
| 2.32e-15 | 211 | 412 | 155 | 365 | Mannan polymerase complex subunit MNN9 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=MNN9 PE=3 SV=1 |
|
| 4.49e-15 | 147 | 413 | 97 | 372 | Mannan polymerase II complex anp1 subunit OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=anp1 PE=3 SV=1 |
|
| 5.02e-15 | 179 | 413 | 138 | 390 | Mannan polymerase complexes subunit MNN9 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN9 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
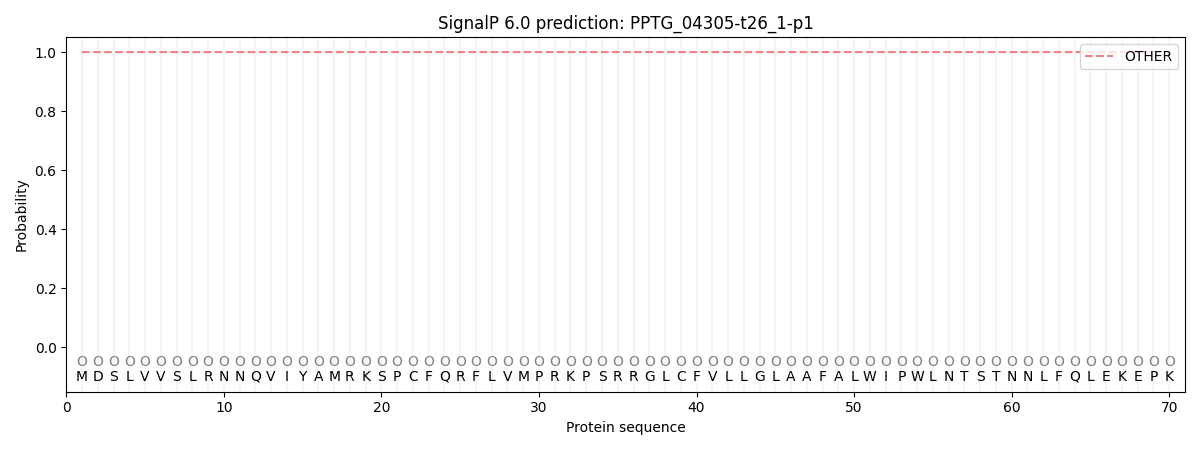
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999983 | 0.000038 |