You are browsing environment: FUNGIDB
CAZyme Information: PPTG_03559-t26_1-p1
You are here: Home > Sequence: PPTG_03559-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_03559-t26_1-p1 | |||||||||||
| CAZy Family | AA6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 44 | 293 | 8.7e-55 | 0.9285714285714286 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396075 | Glyco_hydro_6 | 1.08e-38 | 53 | 290 | 11 | 287 | Glycosyl hydrolases family 6. |
| 227616 | CelA1 | 1.03e-15 | 79 | 347 | 178 | 462 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.55e-83 | 11 | 321 | 5 | 301 | |
| 1.07e-82 | 1 | 344 | 1 | 316 | |
| 2.49e-82 | 35 | 321 | 4 | 301 | |
| 4.73e-78 | 11 | 321 | 5 | 301 | |
| 1.45e-76 | 28 | 330 | 26 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.18e-23 | 88 | 317 | 71 | 291 | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with METHYL-CELLOBIOSYL-4-DEOXY-4-THIO-BETA-D-CELLOBIOSIDE at 1.6 angstrom [Mycobacterium tuberculosis H37Rv] |
|
| 6.18e-23 | 88 | 317 | 71 | 291 | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with cellobiose at 1.75 angstrom [Mycobacterium tuberculosis H37Rv] |
|
| 8.76e-23 | 88 | 317 | 92 | 312 | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with thiocellopentaose at 1.1 angstrom [Mycobacterium tuberculosis H37Rv] |
|
| 1.15e-22 | 88 | 317 | 71 | 291 | Structure of the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with glucose-isofagomine at 1.9 angstrom [Mycobacterium tuberculosis H37Rv] |
|
| 9.17e-21 | 91 | 317 | 79 | 352 | Crystal structure of a fungal chimeric cellobiohydrolase Cel6A [Humicola insolens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.03e-26 | 88 | 288 | 102 | 301 | Endoglucanase A OS=Thermobispora bispora OX=2006 GN=celA PE=3 SV=1 |
|
| 7.87e-20 | 91 | 317 | 181 | 454 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=cbhC PE=3 SV=1 |
|
| 8.29e-20 | 91 | 317 | 190 | 463 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=cbhC PE=3 SV=1 |
|
| 2.42e-19 | 91 | 317 | 172 | 445 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=cbhC PE=3 SV=1 |
|
| 3.34e-19 | 91 | 317 | 176 | 449 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
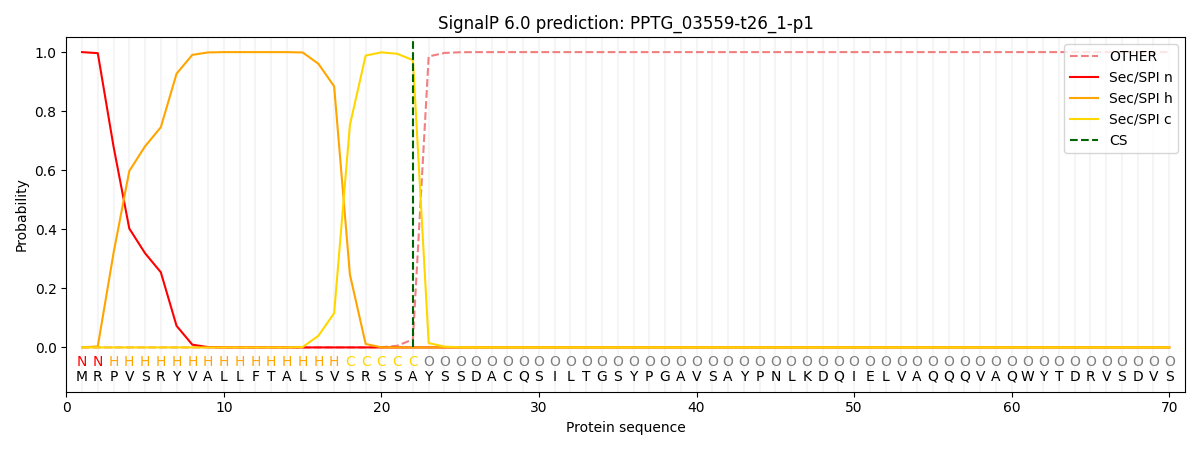
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000241 | 0.999731 | CS pos: 22-23. Pr: 0.9728 |
