You are browsing environment: FUNGIDB
CAZyme Information: PPTG_03153-t26_1-p1
You are here: Home > Sequence: PPTG_03153-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_03153-t26_1-p1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 13 | 261 | 2.1e-75 | 0.9346938775510204 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 235906 | PRK07003 | 5.54e-07 | 189 | 306 | 396 | 515 | DNA polymerase III subunit gamma/tau. |
| 237874 | PRK14971 | 2.09e-06 | 219 | 307 | 386 | 472 | DNA polymerase III subunit gamma/tau. |
| 411474 | fibronec_FbpA | 2.30e-06 | 200 | 305 | 132 | 242 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
| 236090 | PRK07764 | 2.51e-06 | 189 | 298 | 649 | 758 | DNA polymerase III subunits gamma and tau; Validated |
| 236090 | PRK07764 | 3.22e-06 | 203 | 303 | 396 | 494 | DNA polymerase III subunits gamma and tau; Validated |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.09e-166 | 1 | 314 | 1 | 314 | |
| 4.10e-137 | 35 | 314 | 1 | 280 | |
| 1.99e-136 | 35 | 256 | 1 | 222 | |
| 6.66e-106 | 1 | 222 | 13 | 235 | |
| 1.05e-104 | 1 | 223 | 1 | 224 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
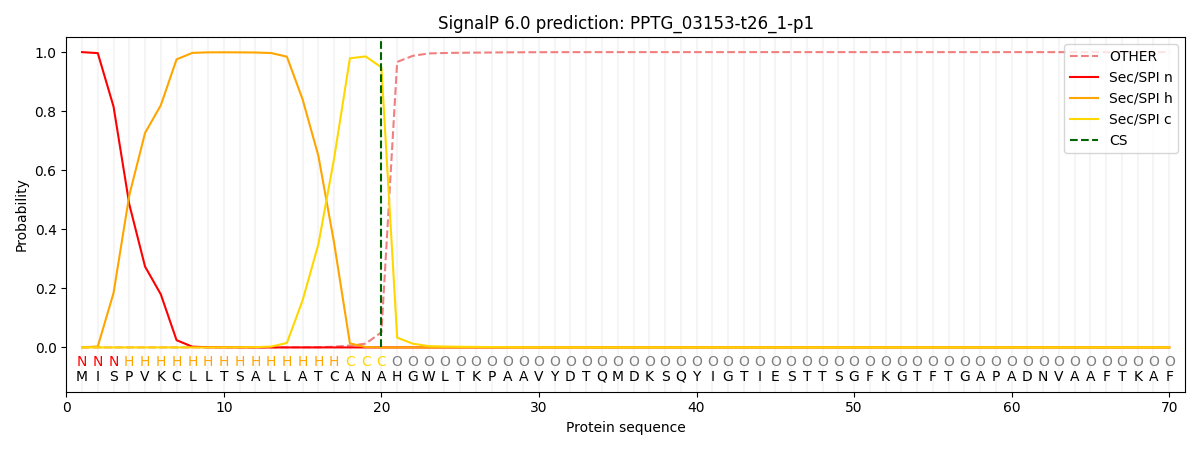
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000796 | 0.999156 | CS pos: 20-21. Pr: 0.9482 |
