You are browsing environment: FUNGIDB
CAZyme Information: PPTG_03139-t26_1-p1
You are here: Home > Sequence: PPTG_03139-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_03139-t26_1-p1 | |||||||||||
| CAZy Family | AA2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 11 | 275 | 1.7e-78 | 0.9755102040816327 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 237030 | kgd | 1.77e-08 | 228 | 311 | 37 | 118 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 237030 | kgd | 5.08e-07 | 225 | 306 | 37 | 118 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 236776 | PRK10856 | 1.37e-06 | 225 | 309 | 148 | 241 | cytoskeleton protein RodZ. |
| 237030 | kgd | 1.65e-06 | 220 | 297 | 40 | 119 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
| 237030 | kgd | 2.50e-06 | 228 | 308 | 34 | 114 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.03e-176 | 1 | 320 | 1 | 320 | |
| 4.18e-176 | 1 | 320 | 1 | 320 | |
| 5.93e-176 | 1 | 320 | 1 | 320 | |
| 8.42e-176 | 1 | 320 | 1 | 320 | |
| 8.42e-176 | 1 | 320 | 1 | 320 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.48e-13 | 24 | 195 | 1 | 170 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
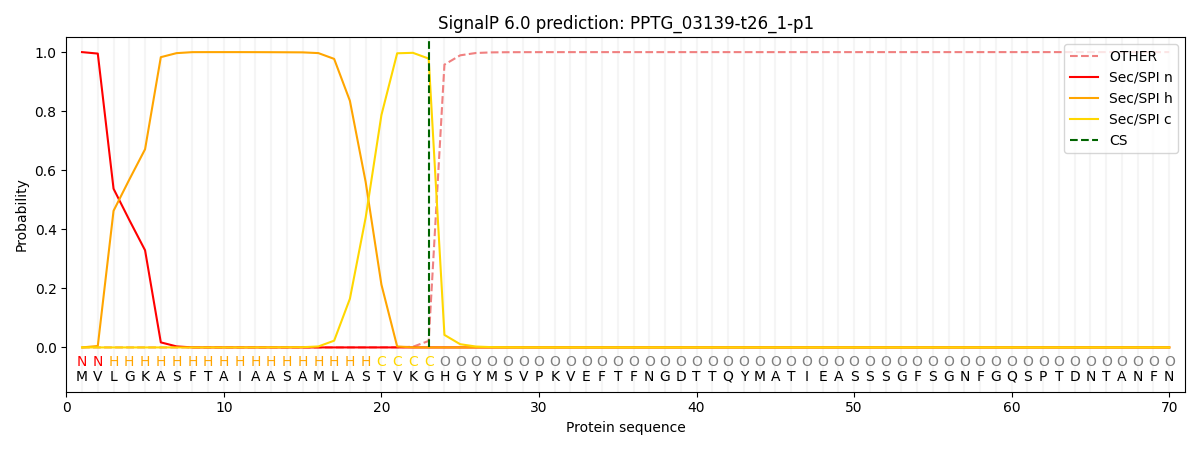
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000261 | 0.999694 | CS pos: 23-24. Pr: 0.9780 |
