You are browsing environment: FUNGIDB
CAZyme Information: PPTG_02510-t26_1-p1
You are here: Home > Sequence: PPTG_02510-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_02510-t26_1-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | hypothetical protein, variant | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1448645; End:1452028 Strand: + | |||||||||||
Full Sequence Download help
| MAAPVWVLCC SVCVAIVAAL SPGFNDPMPY YTLPLLEILH PQNDMMMADT ELQVEIEVRS | 60 |
| ELLSSGLRRS RVCIVMQPAY VPPDVVLDSG AGEVDESCFD QSLNYTTFHV AGLVPGLSYA | 120 |
| LTVGLENDGN MVALSTRTFS VGSILLPGVD DRLSVADALE AGAELHKAQE RETASGIYRY | 180 |
| VLAIFPDHGP AQHLLGLALY QDGKLHEALS LLYRAVQSNE SEENFHNSLG ICLKSLGRIN | 240 |
| EAIKHYRRAI ELNPMQVQAA VNLGDAMQSK GKWEEAFKEY SKVANTPMSL LYSQLEPTKA | 300 |
| EHVAKDATGR LCELIRVTDG WYRADRCLNE ALERWPDEPV FHNDRGNLLA NAGQFETALN | 360 |
| EYQRSSELGL LAGTLNLAET LEALGETQKS IDLYDQVLGS EIFDRFHPRT RIMVMKATVL | 420 |
| PRVLPSSQKE IDAYRDRFER EVEALLHNLD SLETTEVDPN RISLSTAITL TAHNRNNREL | 480 |
| KAAMGQLYWQ LLYRRQLMLE DYVASYGIIP LPYTQQIEDV KRPEIGPQRL RVGFVSRYIF | 540 |
| NSAVGLYMSE LIPKFNRDKY EIVVFAIGQS KSMKVAKEID AVTETIVALP KDMRIVREEI | 600 |
| RAWKMDVLIY PELGMDKTTY FVSLARLAPV QAVWWGNADT SGVPTMDYYL TSEHEHKTAN | 660 |
| SHYSEAIYKF KGMGIYHKLP ALPKRDINRG QVRKAIEERF DISPDFHFYL AIESIIHIHP | 720 |
| DFDAAVAKLF EKDKKAHLFL LSTSSRKIWK SQLQARMESA GVDPDRLHFL TDVDQKQESM | 780 |
| LMRAADAVIA SLHLTRPRAS LQAFAAGVPV VTFPNELWAS RITFGFYQQM GINDLIAASL | 840 |
| DEYVALAVKL ATDSAFHKEM VQLIKRNRSK LSEDEEAVRE WEKFFDFAAD QIFPSGELES | 900 |
| FIPKSESDWG SPQQLKEIQI DWSEAKGLEE WNQAEGWRLR ADEDE | 945 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 310 | 886 | 7.7e-69 | 0.5404255319148936 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 2.44e-30 | 527 | 885 | 257 | 611 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 276809 | TPR | 1.32e-17 | 191 | 283 | 3 | 95 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 9.02e-14 | 159 | 251 | 5 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 411346 | social_mot_Tgl | 3.79e-13 | 195 | 285 | 37 | 128 | social motility TPR repeat lipoprotein Tgl. Social motility in delta-proteobacterial species such as Myxococcus xanthus depends on a type VI pilus, which in turn depends on assembly of the PilQ secretin complex. Tgl, a tetratricopeptide repeat (TPR) outer membrane lipoprotein, is required for PilQ assembly. |
| 223533 | TPR | 7.13e-09 | 156 | 283 | 131 | 262 | Tetratricopeptide (TPR) repeat [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UIZ20641.1|GT41 | 0.0 | 17 | 933 | 19 | 944 |
| BAZ61948.1|GT41 | 1.59e-65 | 333 | 888 | 365 | 884 |
| BAZ15802.1|GT41 | 1.59e-65 | 333 | 888 | 365 | 884 |
| BBO66743.1|GT41 | 5.06e-59 | 414 | 886 | 458 | 899 |
| BBO73735.1|GT41 | 1.75e-58 | 414 | 888 | 374 | 814 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VSN_A | 5.81e-09 | 626 | 855 | 304 | 524 | Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004],2VSN_B Structure and topological arrangement of an O-GlcNAc transferase homolog: insight into molecular control of intracellular glycosylation [Xanthomonas campestris pv. campestris str. 8004] |
| 2JLB_A | 5.81e-09 | 626 | 855 | 304 | 524 | Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2JLB_B Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue [Xanthomonas campestris pv. campestris],2VSY_A Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2VSY_B Xanthomonas campestris putative OGT (XCC0866), apostructure [Xanthomonas campestris pv. campestris str. ATCC 33913],2XGM_A Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGM_B Substrate and product analogues as human O-GlcNAc transferase inhibitors. [Xanthomonas campestris],2XGO_A XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGO_B XcOGT in complex with UDP-S-GlcNAc [Xanthomonas campestris],2XGS_A XcOGT in complex with C-UDP [Xanthomonas campestris],2XGS_B XcOGT in complex with C-UDP [Xanthomonas campestris] |
| 6Q4M_A | 2.36e-07 | 507 | 860 | 218 | 675 | Crystal structure of the O-GlcNAc transferase Asn648Tyr mutation [Homo sapiens] |
| 5HGV_A | 2.10e-06 | 507 | 860 | 214 | 671 | Structure of an O-GlcNAc transferase point mutant, D554N in complex with peptide [Homo sapiens],5HGV_C Structure of an O-GlcNAc transferase point mutant, D554N in complex with peptide [Homo sapiens] |
| 5NPS_A | 2.75e-06 | 507 | 860 | 213 | 670 | The human O-GlcNAc transferase in complex with a bisubstrate inhibitor [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q9M8Y0|SEC_ARATH | 3.84e-10 | 504 | 868 | 568 | 929 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC OS=Arabidopsis thaliana OX=3702 GN=SEC PE=1 SV=1 |
| sp|O82039|SPY_PETHY | 5.09e-08 | 518 | 853 | 479 | 808 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY OS=Petunia hybrida OX=4102 GN=SPY PE=2 SV=1 |
| sp|Q6YZI0|SPY_ORYSJ | 1.51e-07 | 523 | 860 | 467 | 801 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY OS=Oryza sativa subsp. japonica OX=39947 GN=SPY PE=2 SV=1 |
| sp|Q8RVB2|SPY_SOLLC | 2.61e-07 | 527 | 853 | 484 | 808 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY OS=Solanum lycopersicum OX=4081 GN=SPY PE=2 SV=1 |
| sp|O82422|SPY_HORVU | 2.62e-07 | 523 | 860 | 467 | 801 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY OS=Hordeum vulgare OX=4513 GN=SPY PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
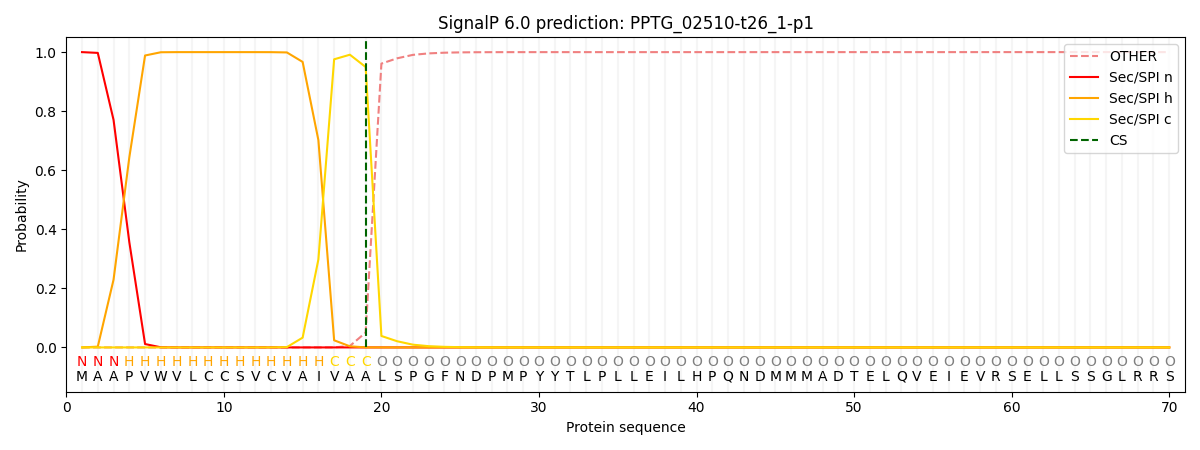
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000217 | 0.999716 | CS pos: 19-20. Pr: 0.9495 |

