You are browsing environment: FUNGIDB
CAZyme Information: PPTG_01153-t26_1-p1
You are here: Home > Sequence: PPTG_01153-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora parasitica | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora parasitica | |||||||||||
| CAZyme ID | PPTG_01153-t26_1-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 9 | 276 | 1.3e-64 | 0.926530612244898 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 183558 | PRK12495 | 2.62e-04 | 211 | 290 | 63 | 145 | hypothetical protein; Provisional |
| 183558 | PRK12495 | 0.001 | 188 | 291 | 76 | 172 | hypothetical protein; Provisional |
| 411196 | act_recrut_TARP | 0.002 | 200 | 289 | 380 | 470 | type III secretion system actin-recruiting effector Tarp. The founding member of the Tarp (translocated actin-recruiting phosphoprotein) is CT456 from Chlamydia trachomatis. Tarp is a type III secretion system effector. Orthologs are found other Chlamydia, but are highly variable in length because many lack much of the repeat region. |
| 236652 | PRK10118 | 0.002 | 185 | 285 | 162 | 262 | flagellar hook length control protein FliK. |
| 411773 | SP4_N | 0.003 | 186 | 280 | 248 | 353 | N-terminal domain of transcription factor Specificity Protein (SP) 4. Specificity Proteins (SPs) are transcription factors that are involved in many cellular processes, including cell differentiation, cell growth, apoptosis, immune responses, response to DNA damage, and chromatin remodeling. Human SP4 is a risk gene of multiple psychiatric disorders including schizophrenia, bipolar disorder, and major depression. SP4 belongs to a family of proteins, called the SP/Kruppel or Krueppel-like Factor (KLF) family, characterized by a C-terminal DNA-binding domain of 81 amino acids consisting of three Kruppel-like C2H2 zinc fingers. These factors bind to a loose consensus motif, namely NNRCRCCYY (where N is any nucleotide; R is A/G, and Y is C/T), such as the recurring motifs in GC and GT boxes (5'-GGGGCGGGG-3' and 5-GGTGTGGGG-3') that are present in promoters and more distal regulatory elements of mammalian genes. SP factors preferentially bind GC boxes, while KLFs bind CACCC boxes. Another characteristic hallmark of SP factors is the presence of the Buttonhead (BTD) box CXCPXC, just N-terminal to the zinc fingers. The function of the BTD box is unknown, but it is thought to play an important physiological role. Another feature of most SP factors is the presence of a conserved amino acid stretch, the so-called SP box, located close to the N-terminus. SP factors may be separated into three groups based on their domain architecture and the similarity of their N-terminal transactivation domains: SP1-4, SP5, and SP6-9. The transactivation domains between the three groups are not homologous to one another. SP1-4 have similar N-terminal transactivation domains characterized by glutamine-rich regions, which, in most cases, have adjacent serine/threonine-rich regions. This model represents the N-terminal domain of SP4. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.23e-164 | 1 | 317 | 1 | 317 | |
| 3.90e-161 | 1 | 317 | 1 | 317 | |
| 4.53e-159 | 1 | 317 | 1 | 303 | |
| 9.14e-159 | 1 | 317 | 1 | 303 | |
| 4.47e-146 | 1 | 317 | 1 | 317 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
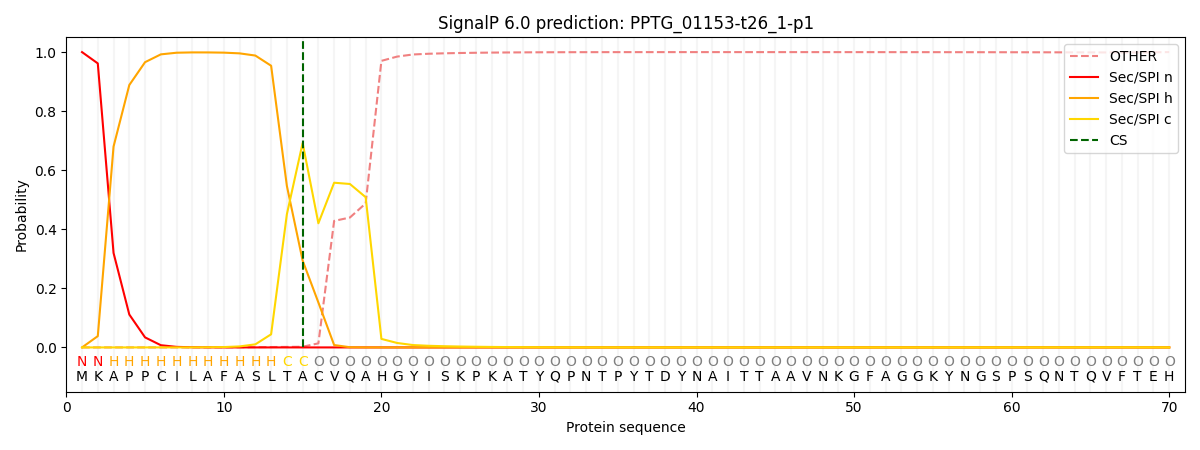
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000591 | 0.999395 | CS pos: 15-16. Pr: 0.6922 |
