You are browsing environment: FUNGIDB
CAZyme Information: POW17911.1
You are here: Home > Sequence: POW17911.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POW17911.1 | |||||||||||
| CAZy Family | GT90 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 46 | 354 | 3.2e-72 | 0.9504950495049505 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395262 | Glyco_hydro_10 | 1.23e-85 | 47 | 356 | 11 | 310 | Glycosyl hydrolase family 10. |
| 214750 | Glyco_10 | 1.33e-76 | 82 | 354 | 1 | 263 | Glycosyl hydrolase family 10. |
| 226217 | XynA | 9.96e-69 | 62 | 359 | 47 | 342 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.02e-76 | 48 | 354 | 37 | 335 | |
| 1.01e-61 | 26 | 354 | 32 | 347 | |
| 1.81e-56 | 47 | 355 | 53 | 353 | |
| 4.84e-56 | 50 | 350 | 49 | 338 | |
| 4.90e-56 | 63 | 355 | 53 | 339 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.94e-50 | 62 | 350 | 32 | 311 | Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum [Gloeophyllum trabeum ATCC 11539],4XX6_B Crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Gloeophyllum trabeum [Gloeophyllum trabeum ATCC 11539] |
|
| 1.61e-48 | 64 | 355 | 33 | 317 | Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBR_B Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose [Thermotoga maritima],1VBU_A Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima],1VBU_B Crystal structure of native xylanase 10B from Thermotoga maritima [Thermotoga maritima] |
|
| 3.13e-48 | 64 | 355 | 49 | 333 | Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NIY_B Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3NJ3_A Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1],3NJ3_B Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose [Thermotoga petrophila RKU-1] |
|
| 2.75e-44 | 64 | 358 | 28 | 312 | Beta-1,4-Glycanase Cex-Cd [Cellulomonas fimi],1FH7_A Crystal Structure Of The Xylanase Cex With Xylobiose- Derived Inhibitor Deoxynojirimycin [Cellulomonas fimi],1FH8_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Isofagomine Inhibitor [Cellulomonas fimi],1FH9_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Lactam Oxime Inhibitor [Cellulomonas fimi],1FHD_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Imidazole Inhibitor [Cellulomonas fimi],1J01_A Crystal Structure Of The Xylanase Cex With Xylobiose-Derived Inhibitor Isofagomine lactam [Cellulomonas fimi],2EXO_A Crystal Structure Of The Catalytic Domain Of The Beta-1,4- Glycanase Cex From Cellulomonas Fimi [Cellulomonas fimi],2XYL_A Cellulomonas Fimi XylanaseCELLULASE COMPLEXED WITH 2-Deoxy- 2-Fluoro-Xylobiose [Cellulomonas fimi] |
|
| 2.96e-44 | 64 | 358 | 28 | 312 | Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellobiose-like isofagomine [Cellulomonas fimi],3CUG_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellotetraose-like isofagomine [Cellulomonas fimi],3CUH_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellotriose-like isofagomine [Cellulomonas fimi],3CUI_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with sulfur substituted beta-1,4 xylotetraose [Cellulomonas fimi],3CUJ_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with sulfur substituted beta-1,4 xylopentaose. [Cellulomonas fimi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.29e-43 | 64 | 347 | 52 | 328 | Endo-1,4-beta-xylanase B OS=Thermotoga neapolitana OX=2337 GN=xynB PE=3 SV=1 |
|
| 1.27e-42 | 51 | 354 | 32 | 327 | Endo-1,4-beta-xylanase OS=Agaricus bisporus OX=5341 GN=xlnA PE=2 SV=1 |
|
| 4.84e-41 | 64 | 358 | 69 | 353 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
|
| 6.74e-41 | 64 | 354 | 67 | 346 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
|
| 8.08e-38 | 62 | 355 | 542 | 831 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
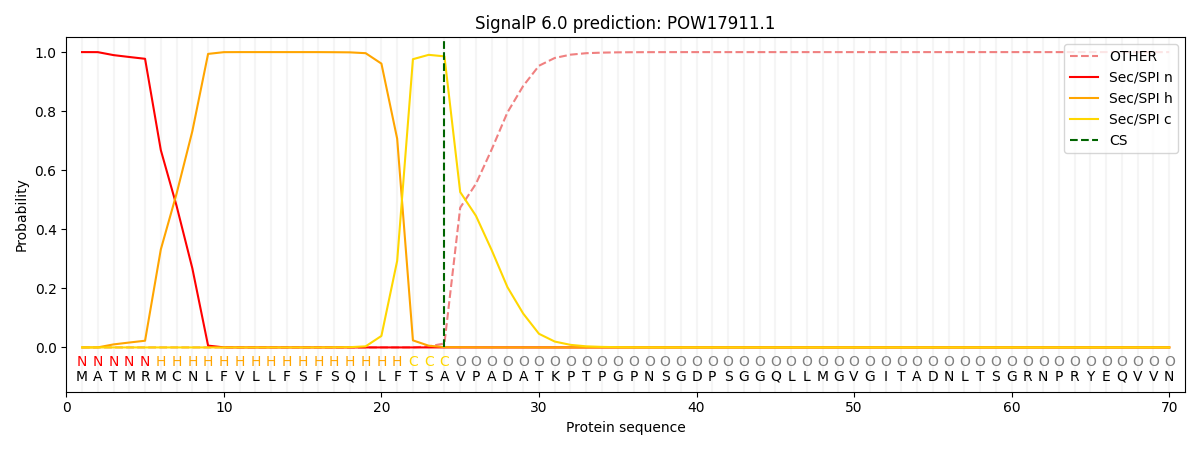
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000198 | 0.999785 | CS pos: 24-25. Pr: 0.9856 |
