You are browsing environment: FUNGIDB
CAZyme Information: POW14050.1
You are here: Home > Sequence: POW14050.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POW14050.1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | Glucanase [Source:UniProtKB/TrEMBL;Acc:A0A2S4W6B1] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.176:83 | 3.2.1.132:6 | 3.2.1.4:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH7 | 27 | 444 | 4.8e-129 | 0.9783132530120482 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395677 | Glyco_hydro_7 | 0.0 | 22 | 444 | 1 | 433 | Glycosyl hydrolase family 7. |
| 153432 | GH7_CBH_EG | 7.05e-128 | 29 | 439 | 2 | 385 | Glycosyl hydrolase family 7. Glycosyl hydrolase family 7 contains eukaryotic endoglucanases (EGs) and cellobiohydrolases (CBHs) that hydrolyze glycosidic bonds using a double-displacement mechanism. This leads to a net retention of the conformation at the anomeric carbon. Both enzymes work synergistically in the degradation of cellulose,which is the main component of plant cell wall, and is composed of beta-1,4 linked glycosyl units. EG cleaves the beta-1,4 linkages of cellulose and CBH cleaves off cellobiose disaccharide units from the reducing end of the chain. In general, the O-glycosyl hydrolases are a widespread group of enzymes that hydrolyze the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A glycosyl hydrolase classification system based on sequence similarity has led to the definition of more than 95 different families inlcuding glycoside hydrolase family 7. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.78e-116 | 18 | 444 | 17 | 454 | |
| 1.21e-114 | 18 | 444 | 17 | 454 | |
| 2.52e-110 | 16 | 444 | 14 | 450 | |
| 5.05e-110 | 8 | 444 | 7 | 452 | |
| 6.59e-110 | 17 | 444 | 16 | 456 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.63e-107 | 29 | 444 | 9 | 434 | Dictyostelium purpureum cellobiohydrolase Cel7A apo structure [Dictyostelium purpureum],4ZZP_B Dictyostelium purpureum cellobiohydrolase Cel7A apo structure [Dictyostelium purpureum] |
|
| 1.41e-103 | 29 | 444 | 9 | 436 | Structure of Cellobiohydrolase 1 (Cel7A) from Heterobasidion annosum [Heterobasidion annosum],2YG1_A APO STRUCTURE OF CELLOBIOHYDROLASE 1 (CEL7A) FROM HETEROBASIDION ANNOSUM [Heterobasidion annosum],2YG1_B APO STRUCTURE OF CELLOBIOHYDROLASE 1 (CEL7A) FROM HETEROBASIDION ANNOSUM [Heterobasidion annosum] |
|
| 1.12e-102 | 29 | 444 | 9 | 435 | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus [Aspergillus fumigatus],4V20_A The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus, disaccharide complex [Aspergillus fumigatus] |
|
| 6.77e-102 | 29 | 444 | 9 | 425 | Chain A, EXOGLUCANASE I [Phanerodontia chrysosporium],1H46_X Chain X, EXOGLUCANASE I [Phanerodontia chrysosporium],1Z3T_A Chain A, cellulase [Phanerodontia chrysosporium],1Z3V_A Chain A, cellulase [Phanerodontia chrysosporium],1Z3W_A Chain A, cellulase [Phanerodontia chrysosporium] |
|
| 2.29e-101 | 29 | 444 | 9 | 432 | Chain A, cellobiohydrolase I catalytic domain [Rasamsonia emersonii],3PFJ_A Chain A, Cellobiohydrolase 1 catalytic domain [Rasamsonia emersonii],3PFX_A Chain A, Cellobiohydrolase 1 catalytic domain [Rasamsonia emersonii],3PFZ_A Chain A, Cellobiohydrolase 1 catalytic domain [Rasamsonia emersonii],3PL3_A Chain A, Cellobiohydrolase 1 catalytic domain [Rasamsonia emersonii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.90e-107 | 19 | 444 | 26 | 461 | Probable 1,4-beta-D-glucan cellobiohydrolase B OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=cbhB PE=3 SV=1 |
|
| 1.13e-106 | 19 | 444 | 22 | 459 | 1,4-beta-D-glucan cellobiohydrolase B OS=Aspergillus aculeatus OX=5053 GN=cbhB PE=2 SV=1 |
|
| 1.18e-106 | 8 | 444 | 7 | 448 | Exoglucanase OS=Agaricus bisporus OX=5341 GN=cel2 PE=2 SV=1 |
|
| 1.98e-104 | 20 | 444 | 19 | 443 | Exoglucanase 1 OS=Phanerodontia chrysosporium OX=2822231 GN=CBH1 PE=2 SV=1 |
|
| 2.32e-102 | 19 | 444 | 23 | 458 | 1,4-beta-D-glucan cellobiohydrolase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cbhB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
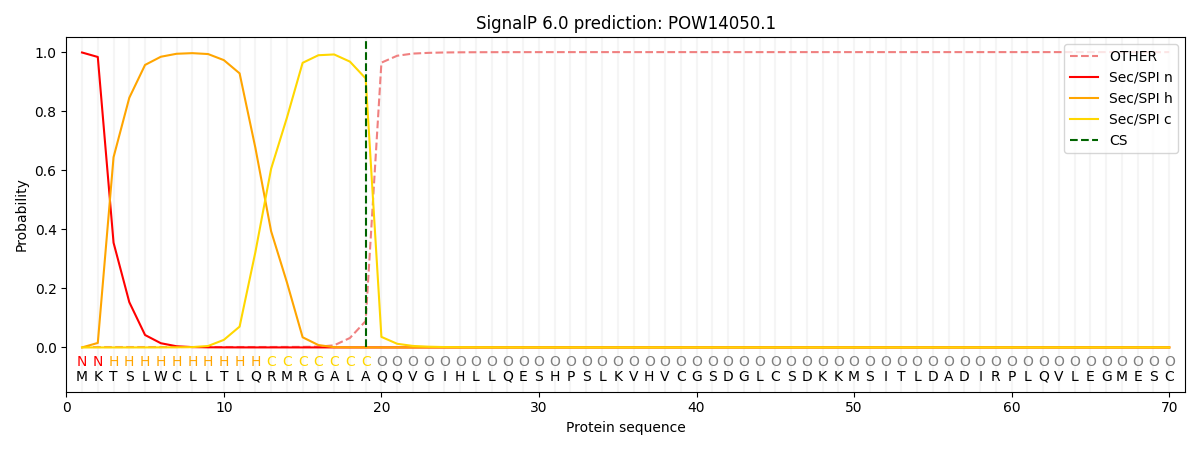
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.002851 | 0.997125 | CS pos: 19-20. Pr: 0.9108 |
