You are browsing environment: FUNGIDB
CAZyme Information: POW13688.1
You are here: Home > Sequence: POW13688.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
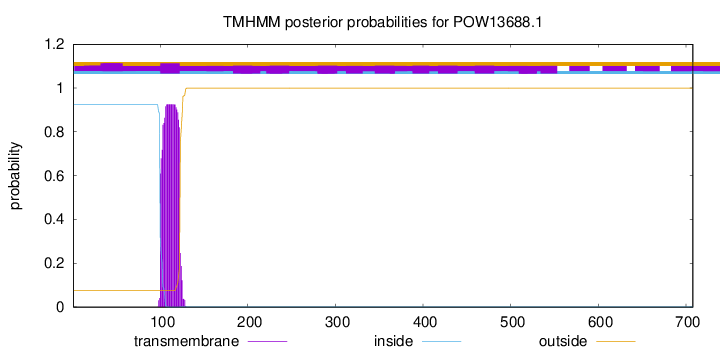
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POW13688.1 | |||||||||||
| CAZy Family | GT10 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214773 | CAP10 | 2.74e-13 | 571 | 685 | 128 | 241 | Putative lipopolysaccharide-modifying enzyme. |
| 310354 | Glyco_transf_90 | 1.30e-09 | 586 | 695 | 213 | 321 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.98e-156 | 102 | 706 | 153 | 752 | |
| 7.72e-110 | 176 | 707 | 169 | 703 | |
| 1.34e-102 | 176 | 702 | 114 | 637 | |
| 2.59e-102 | 176 | 706 | 171 | 694 | |
| 5.09e-100 | 169 | 708 | 167 | 711 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.00e-10 | 542 | 698 | 196 | 343 | human POGLUT1 in complex with Notch1 EGF12 and UDP [Homo sapiens],5L0S_A human POGLUT1 in complex with Factor VII EGF1 and UDP [Homo sapiens],5L0T_A human POGLUT1 in complex with EGF(+) and UDP [Homo sapiens],5L0U_A human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose [Homo sapiens],5L0V_A human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP [Homo sapiens],5UB5_A human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP [Homo sapiens] |
|
| 2.04e-09 | 578 | 692 | 261 | 373 | Crystal structure of Drosophila Poglut1 (Rumi) complexed with its glycoprotein product (glucosylated EGF repeat) and UDP [Drosophila melanogaster],5F85_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) and UDP [Drosophila melanogaster],5F86_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with its substrate protein (EGF repeat) [Drosophila melanogaster],5F87_A Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_B Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_C Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_D Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_E Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster],5F87_F Crystal structure of Drosophila Poglut1 (Rumi) complexed with UDP [Drosophila melanogaster] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.39e-81 | 171 | 703 | 147 | 662 | Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
|
| 7.89e-10 | 542 | 698 | 224 | 371 | Protein O-glucosyltransferase 1 OS=Rattus norvegicus OX=10116 GN=Poglut1 PE=3 SV=1 |
|
| 2.45e-09 | 542 | 698 | 224 | 371 | Protein O-glucosyltransferase 1 OS=Bos taurus OX=9913 GN=POGLUT1 PE=2 SV=1 |
|
| 2.45e-09 | 542 | 698 | 224 | 371 | Protein O-glucosyltransferase 1 OS=Homo sapiens OX=9606 GN=POGLUT1 PE=1 SV=1 |
|
| 1.01e-08 | 542 | 685 | 224 | 358 | Protein O-glucosyltransferase 1 OS=Mus musculus OX=10090 GN=Poglut1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000041 | 0.000000 |