You are browsing environment: FUNGIDB
CAZyme Information: POW11743.1
You are here: Home > Sequence: POW11743.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
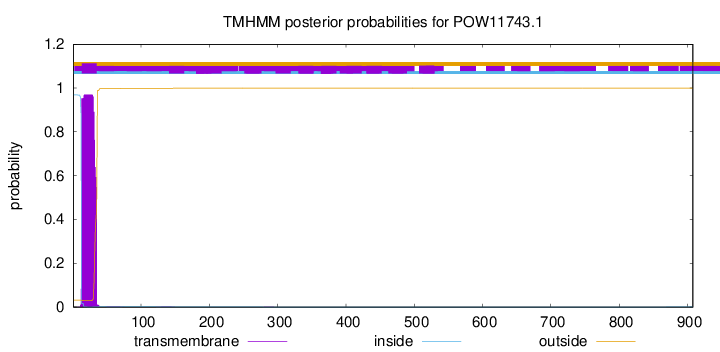
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POW11743.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.22:58 | 2.4.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 202 | 371 | 1.3e-28 | 0.759825327510917 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 399040 | TANGO2 | 2.15e-41 | 561 | 871 | 6 | 250 | Transport and Golgi organisation 2. In eukaryotes this family is predicted to play a role in protein secretion and Golgi organisation. In plants this family includes Solanum habrochaites Cwp, which is involved in water permeability in the cuticles of fruit. Mouse Tango2 has been found to be expressed during early embryogenesis in mice. This protein contains a conserved NRDE motif. This gene has been characterized in Drosophila melanogaster and named as transport and Golgi organisation 2, hence the name Tango2. |
| 269893 | GH27 | 3.90e-36 | 60 | 318 | 2 | 271 | glycosyl hydrolase family 27 (GH27). GH27 enzymes occur in eukaryotes, prokaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-N-acetylgalactosaminidase, and 3-alpha-isomalto-dextranase. All GH27 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH27 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| 166449 | PLN02808 | 1.20e-27 | 61 | 394 | 34 | 384 | alpha-galactosidase |
| 178295 | PLN02692 | 1.92e-26 | 61 | 338 | 58 | 341 | alpha-galactosidase |
| 177874 | PLN02229 | 1.15e-17 | 61 | 388 | 65 | 412 | alpha-galactosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.72e-53 | 60 | 433 | 30 | 440 | |
| 2.72e-53 | 60 | 433 | 30 | 440 | |
| 2.72e-53 | 60 | 433 | 30 | 440 | |
| 1.30e-51 | 60 | 404 | 28 | 401 | |
| 1.73e-28 | 61 | 394 | 54 | 404 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.66e-26 | 61 | 394 | 11 | 360 | Chain A, alpha-galactosidase [Oryza sativa] |
|
| 5.58e-26 | 61 | 394 | 11 | 361 | Nicotiana benthamiana alpha-galactosidase [Nicotiana benthamiana] |
|
| 2.87e-18 | 58 | 393 | 11 | 410 | Chain A, alpha-galactosidase [Trichoderma reesei],1T0O_A Chain A, alpha-galactosidase [Trichoderma reesei] |
|
| 2.43e-17 | 61 | 392 | 11 | 389 | Crystal structure of alpha-galactosidase I from Mortierella vinacea [Umbelopsis vinacea] |
|
| 3.45e-15 | 59 | 454 | 12 | 439 | Crystal Structure of Streptomyces avermitilis beta-L-Arabinopyranosidase [Streptomyces avermitilis],3A21_B Crystal Structure of Streptomyces avermitilis beta-L-Arabinopyranosidase [Streptomyces avermitilis],3A22_A Crystal Structure of beta-L-Arabinopyranosidase complexed with L-arabinose [Streptomyces avermitilis],3A22_B Crystal Structure of beta-L-Arabinopyranosidase complexed with L-arabinose [Streptomyces avermitilis],3A23_A Crystal Structure of beta-L-Arabinopyranosidase complexed with D-galactose [Streptomyces avermitilis],3A23_B Crystal Structure of beta-L-Arabinopyranosidase complexed with D-galactose [Streptomyces avermitilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.98e-25 | 61 | 394 | 66 | 415 | Alpha-galactosidase OS=Oryza sativa subsp. japonica OX=39947 GN=Os10g0493600 PE=1 SV=1 |
|
| 1.25e-23 | 61 | 397 | 42 | 395 | Alpha-galactosidase 2 OS=Arabidopsis thaliana OX=3702 GN=AGAL2 PE=1 SV=1 |
|
| 1.28e-23 | 61 | 399 | 36 | 403 | Probable alpha-galactosidase D OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=aglD PE=3 SV=2 |
|
| 1.72e-23 | 61 | 395 | 35 | 401 | Alpha-galactosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=aglD PE=1 SV=2 |
|
| 7.01e-23 | 61 | 393 | 34 | 401 | Probable alpha-galactosidase D OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=aglD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000031 | 0.000001 |