You are browsing environment: FUNGIDB
CAZyme Information: POW03045.1
You are here: Home > Sequence: POW03045.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
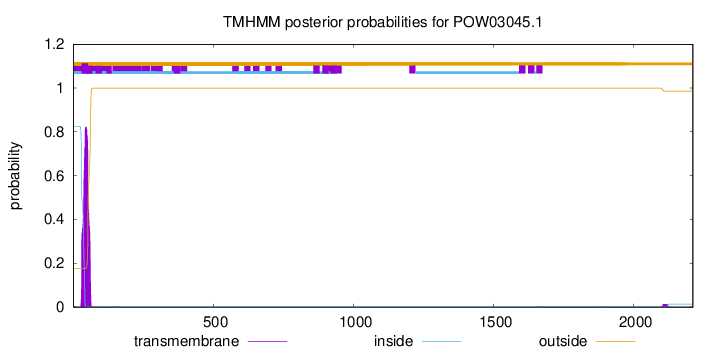
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POW03045.1 | |||||||||||
| CAZy Family | GH131 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA1 | 165 | 672 | 2.2e-50 | 0.7318435754189944 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 224557 | HrpA | 1.09e-142 | 1344 | 2069 | 7 | 657 | HrpA-like RNA helicase [Translation, ribosomal structure and biogenesis]. |
| 259950 | CuRO_2_Diphenol_Ox | 3.79e-91 | 199 | 365 | 1 | 164 | The second cupredoxin domain of fungal laccase, diphenol oxidase. Diphenol oxidase belongs to the laccase family. It catalyzes the initial steps in melanin biosynthesis from diphenols. Melanin is one of the virulence factors of infectious fungi. In the pathogenesis of C. neoformans, melanin pigments have been shown to protect the fungal cells from oxidative and microbicidal activities of host defense systems. Laccase is a blue multi-copper enzyme that catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. Laccase is a multicopper oxidase (MCO) composed of three cupredoxin domains that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 2 of 3-domain MCOs has lost the ability to bind copper. |
| 259971 | CuRO_3_Diphenol_Ox | 3.63e-75 | 533 | 680 | 2 | 153 | The third cupredoxin domain of fungal laccase, diphenol oxidase. Diphenol oxidase belongs to the laccase family. It catalyzes the initial steps in melanin biosynthesis from diphenols. Melanin is one of the virulence factors of infectious fungi. In the pathogenesis of C. neoformans, melanin pigments have been shown to protect the fungal cells from oxidative and microbicidal activities of host defense systems. Laccase is a blue multicopper oxidase (MCO) which catalyzes the oxidation of a variety aromatic - notably phenolic and inorganic substances coupled to the reduction of molecular oxygen to water. It has been implicated in a wide spectrum of biological activities and, in particular, plays a key role in morphogenesis, development and lignin metabolism. Although MCOs have diverse functions, majority of them have three cupredoxin domain repeats that include one mononuclear and one trinuclear copper center. The copper ions are bound in several sites: Type 1, Type 2, and/or Type 3. The ensemble of types 2 and 3 copper is called a trinuclear cluster. MCOs oxidize their substrate by accepting electrons at a mononuclear copper center and transferring them to the active site trinuclear copper center. The cupredoxin domain 3 of 3-domain MCOs contains the Type 1 (T1) copper binding site and part the trinuclear copper binding site, which is located at the interface of domains 1 and 3. |
| 182986 | PRK11131 | 2.60e-71 | 1375 | 1958 | 71 | 589 | ATP-dependent RNA helicase HrpA; Provisional |
| 350675 | DEXHc_RHA-like | 2.77e-64 | 1396 | 1559 | 2 | 159 | DEXH-box helicase domain of DEAD-like helicase RHA family proteins. The RNA helicase A (RHA) family includes RHA, also called DEAH-box helicase 9 (DHX9), DHX8, DHX15-16, DHX32-38, and many others. The RHA family belongs to the DEAD-like helicase superfamily, a diverse family of proteins involved in ATP-dependent RNA or DNA unwinding. This domain contains the ATP-binding region. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.62e-121 | 72 | 706 | 50 | 599 | |
| 8.60e-119 | 72 | 706 | 50 | 599 | |
| 1.15e-114 | 81 | 749 | 39 | 599 | |
| 1.15e-114 | 81 | 749 | 39 | 599 | |
| 1.81e-111 | 81 | 749 | 39 | 599 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.76e-107 | 1351 | 2204 | 22 | 830 | Crystal structure of the murine DHX36 helicase [Mus musculus],6UP3_A Crystal structure of the murine DHX36 helicase in complex with ANP [Mus musculus],6UP4_A Crystal structure of the murine DHX36 helicase in complex with ADP [Mus musculus] |
|
| 3.13e-106 | 1351 | 2204 | 34 | 842 | DHX36 with an N-terminal truncation [Bos taurus],5VHC_D DHX36 with an N-terminal truncation bound to ADP-BeF3 [Bos taurus],5VHD_D DHX36 with an N-terminal truncation bound to ADP-AlF4 [Bos taurus] |
|
| 9.31e-106 | 1351 | 2204 | 97 | 905 | DHX36 in complex with the c-Myc G-quadruplex [Bos taurus] |
|
| 6.62e-92 | 1348 | 2118 | 124 | 855 | Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC [Drosophila melanogaster],5N8R_B Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC [Drosophila melanogaster],5N8S_A Crystal Structure of Drosophila DHX36 helicase in complex with polyT [Drosophila melanogaster],5N8S_B Crystal Structure of Drosophila DHX36 helicase in complex with polyT [Drosophila melanogaster],5N8U_A Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCT [Drosophila melanogaster],5N8U_B Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCT [Drosophila melanogaster],5N8Z_A Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCCTT [Drosophila melanogaster],5N8Z_B Crystal Structure of Drosophila DHX36 helicase in complex with CTCTCCCTT [Drosophila melanogaster],5N90_A Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT [Drosophila melanogaster],5N90_B Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT [Drosophila melanogaster],5N94_A Crystal Structure of Drosophila DHX36 helicase in complex with polyU [Drosophila melanogaster],5N94_B Crystal Structure of Drosophila DHX36 helicase in complex with polyU [Drosophila melanogaster],5N96_A Crystal Structure of Drosophila DHX36 helicase in complex with AGGGTTTTTT [Drosophila melanogaster],5N96_B Crystal Structure of Drosophila DHX36 helicase in complex with AGGGTTTTTT [Drosophila melanogaster],5N98_A Crystal Structure of Drosophila DHX36 helicase in complex with TAGGGTTTT [Drosophila melanogaster],5N98_B Crystal Structure of Drosophila DHX36 helicase in complex with TAGGGTTTT [Drosophila melanogaster],5N9A_A Crystal Structure of Drosophila DHX36 helicase in complex with GTTAGGGTT [Drosophila melanogaster],5N9A_B Crystal Structure of Drosophila DHX36 helicase in complex with GTTAGGGTT [Drosophila melanogaster],5N9D_A Crystal Structure of Drosophila DHX36 helicase in complex with GGGTTAGGGT [Drosophila melanogaster],5N9D_B Crystal Structure of Drosophila DHX36 helicase in complex with GGGTTAGGGT [Drosophila melanogaster],5N9E_A Crystal Structure of Drosophila DHX36 helicase in complex with TGGGGATTT [Drosophila melanogaster],5N9E_B Crystal Structure of Drosophila DHX36 helicase in complex with TGGGGATTT [Drosophila melanogaster],5N9F_A Crystal Structure of Drosophila DHX36 helicase in complex with ssDNA CpG_A [Drosophila melanogaster],5N9F_B Crystal Structure of Drosophila DHX36 helicase in complex with ssDNA CpG_A [Drosophila melanogaster] |
|
| 1.29e-77 | 1367 | 1975 | 374 | 976 | Structure of MLE RNA ADP AlF4 complex [Drosophila melanogaster],5AOR_B Structure of MLE RNA ADP AlF4 complex [Drosophila melanogaster] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 776 | 2208 | 89 | 1566 | Putative DEAH-box ATP-dependent helicase UM11114 OS=Ustilago maydis (strain 521 / FGSC 9021) OX=237631 GN=UMAG_11114 PE=3 SV=1 |
|
| 2.19e-146 | 782 | 2207 | 66 | 1327 | Putative ATP-dependent RNA helicase ucp12 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ucp12 PE=3 SV=1 |
|
| 2.27e-119 | 1354 | 2204 | 205 | 992 | DExH-box ATP-dependent RNA helicase DExH1 OS=Arabidopsis thaliana OX=3702 GN=At2g35920 PE=2 SV=1 |
|
| 3.89e-118 | 782 | 2205 | 61 | 1434 | Putative ATP-dependent RNA helicase YLR419W OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YLR419W PE=1 SV=1 |
|
| 4.53e-113 | 1363 | 2126 | 531 | 1321 | Putative ATP-dependent RNA helicase DHX57 OS=Mus musculus OX=10090 GN=Dhx57 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.998739 | 0.001255 |