You are browsing environment: FUNGIDB
CAZyme Information: POW02844.1
You are here: Home > Sequence: POW02844.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
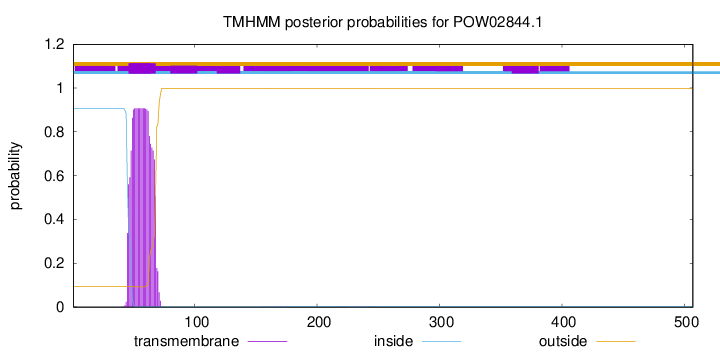
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POW02844.1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226444 | COG3934 | 1.00e-05 | 280 | 409 | 95 | 209 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.04e-204 | 123 | 506 | 85 | 475 | |
| 7.18e-116 | 169 | 507 | 224 | 570 | |
| 1.05e-92 | 161 | 507 | 144 | 503 | |
| 5.99e-88 | 150 | 507 | 157 | 529 | |
| 6.47e-88 | 158 | 507 | 115 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.23e-24 | 175 | 409 | 6 | 254 | Chain A, endo-beta-mannanase [Solanum lycopersicum] |
|
| 2.13e-23 | 257 | 434 | 96 | 308 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
|
| 1.01e-22 | 257 | 434 | 82 | 294 | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
|
| 9.73e-20 | 174 | 434 | 3 | 294 | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
|
| 8.69e-14 | 175 | 411 | 24 | 288 | Exo-mannosidase from Cellvibrio mixtus [Cellvibrio mixtus],1UZ4_A Common inhibition of beta-glucosidase and beta-mannosidase by isofagomine lactam reflects different conformational intineraries for glucoside and mannoside hydrolysis [Cellvibrio mixtus],7ODJ_AAA Chain AAA, Man5A [Cellvibrio mixtus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.65e-29 | 169 | 385 | 31 | 250 | Mannan endo-1,4-beta-mannosidase 1 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN1 PE=2 SV=2 |
|
| 8.56e-27 | 175 | 503 | 31 | 401 | Mannan endo-1,4-beta-mannosidase 7 OS=Arabidopsis thaliana OX=3702 GN=MAN7 PE=2 SV=1 |
|
| 2.19e-26 | 165 | 484 | 12 | 378 | Mannan endo-1,4-beta-mannosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN4 PE=2 SV=2 |
|
| 7.13e-26 | 164 | 385 | 73 | 289 | Putative mannan endo-1,4-beta-mannosidase 5 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN5 PE=2 SV=2 |
|
| 5.19e-25 | 175 | 419 | 54 | 316 | Mannan endo-1,4-beta-mannosidase 2 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN2 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.952994 | 0.046994 |