You are browsing environment: FUNGIDB
CAZyme Information: POV97749.1
You are here: Home > Sequence: POV97749.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
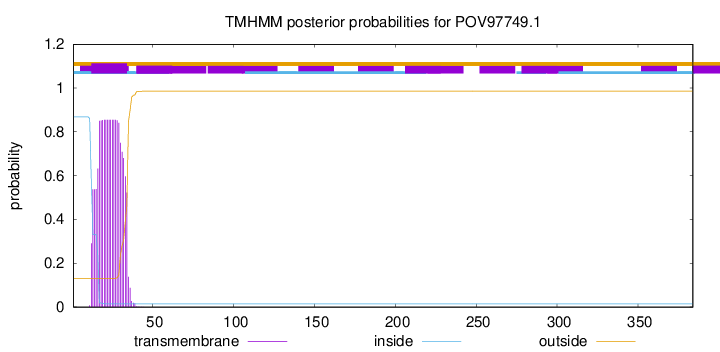
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POV97749.1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.214:21 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT10 | 76 | 363 | 4e-50 | 0.7492795389048992 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395683 | Glyco_transf_10 | 3.20e-29 | 171 | 360 | 2 | 162 | Glycosyltransferase family 10 (fucosyltransferase) C-term. This is the C-terminal domain of a family of fucosyltransferases. This enzyme transfers fucose from GDP-Fucose to GlcNAc in an alpha1,3 linkage. This family is known as glycosyltransferase family 10. The C-terminal domain is the likely binding-region for ADP (manuscript in publication). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.65e-54 | 39 | 332 | 92 | 412 | |
| 3.76e-36 | 100 | 357 | 157 | 381 | |
| 3.76e-36 | 100 | 357 | 157 | 381 | |
| 3.76e-36 | 100 | 357 | 157 | 381 | |
| 3.76e-36 | 100 | 357 | 157 | 381 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.07e-06 | 239 | 327 | 223 | 308 | Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_B Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZW_C Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],2NZX_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZX_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP [Helicobacter pylori],2NZY_A Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_B Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori],2NZY_C Crystal Structure of alpha1,3-Fucosyltransferase with GDP-fucose [Helicobacter pylori] |
|
| 1.19e-06 | 239 | 327 | 223 | 308 | Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori],5ZOI_B Crystal Structure of alpha1,3-Fucosyltransferase [Helicobacter pylori] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.67e-37 | 100 | 357 | 157 | 381 | Putative fucosyltransferase-like protein OS=Arabidopsis thaliana OX=3702 GN=FUT12 PE=2 SV=1 |
|
| 1.60e-32 | 101 | 357 | 147 | 370 | Glycoprotein 3-alpha-L-fucosyltransferase A OS=Arabidopsis thaliana OX=3702 GN=FUT11 PE=1 SV=1 |
|
| 3.39e-17 | 135 | 332 | 152 | 321 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9 OS=Rattus norvegicus OX=10116 GN=Fut9 PE=1 SV=1 |
|
| 8.40e-17 | 135 | 332 | 152 | 321 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9 OS=Mus musculus OX=10090 GN=Fut9 PE=1 SV=1 |
|
| 1.54e-16 | 135 | 332 | 152 | 321 | 4-galactosyl-N-acetylglucosaminide 3-alpha-L-fucosyltransferase 9 OS=Bos taurus OX=9913 GN=FUT9 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001508 | 0.998452 | CS pos: 33-34. Pr: 0.9557 |