You are browsing environment: FUNGIDB
CAZyme Information: POV97020.1
You are here: Home > Sequence: POV97020.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Puccinia striiformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia striiformis | |||||||||||
| CAZyme ID | POV97020.1 | |||||||||||
| CAZy Family | AA6 | |||||||||||
| CAZyme Description | Glyco_trans_2-like domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A2S4UID9] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 527 | 725 | 2.5e-26 | 0.9898477157360406 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404513 | Glyco_trans_2_3 | 2.99e-22 | 527 | 726 | 1 | 194 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 404520 | Glyco_tranf_2_3 | 2.81e-06 | 525 | 659 | 88 | 230 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
| 224136 | BcsA | 3.59e-05 | 461 | 729 | 71 | 348 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 224748 | OafA | 8.42e-04 | 190 | 303 | 254 | 362 | Peptidoglycan/LPS O-acetylase OafA/YrhL, contains acyltransferase and SGNH-hydrolase domains [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 21 | 778 | 380 | 1230 | |
| 5.92e-176 | 23 | 776 | 203 | 1069 | |
| 1.87e-173 | 24 | 776 | 202 | 1067 | |
| 4.04e-172 | 24 | 776 | 202 | 1067 | |
| 1.13e-171 | 24 | 776 | 202 | 1067 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.47e-31 | 422 | 781 | 56 | 453 | Beta-1,4-mannosyltransferase egh OS=Drosophila melanogaster OX=7227 GN=egh PE=2 SV=1 |
|
| 5.84e-26 | 422 | 660 | 56 | 330 | Beta-1,4-mannosyltransferase bre-3 OS=Caenorhabditis briggsae OX=6238 GN=bre-3 PE=3 SV=1 |
|
| 1.41e-25 | 422 | 660 | 56 | 330 | Beta-1,4-mannosyltransferase bre-3 OS=Caenorhabditis elegans OX=6239 GN=bre-3 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999972 | 0.000044 |
TMHMM Annotations download full data without filtering help
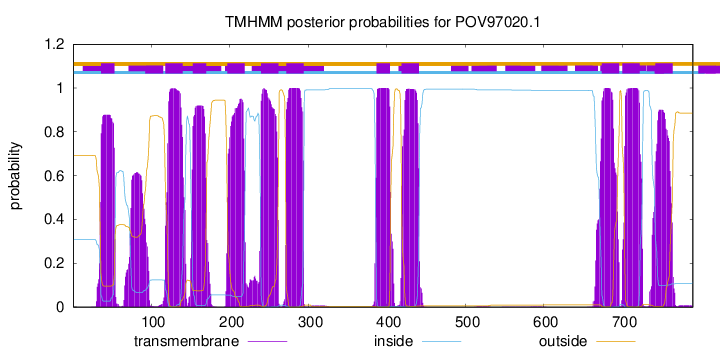
| Start | End |
|---|---|
| 36 | 53 |
| 118 | 140 |
| 153 | 170 |
| 197 | 219 |
| 240 | 262 |
| 272 | 294 |
| 387 | 404 |
| 419 | 441 |
| 674 | 696 |
| 700 | 722 |
| 742 | 764 |
