You are browsing environment: FUNGIDB
CAZyme Information: PNEJI1_003151-t26_1-p1
You are here: Home > Sequence: PNEJI1_003151-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
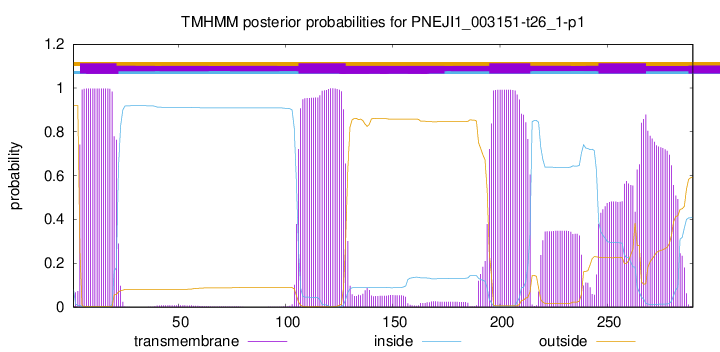
TMHMM annotations
Basic Information help
| Species | Pneumocystis jirovecii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis jirovecii | |||||||||||
| CAZyme ID | PNEJI1_003151-t26_1-p1 | |||||||||||
| CAZy Family | GT44 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.142:10 | 2.4.1.-:1 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340843 | GT33_ALG1-like | 3.28e-61 | 37 | 219 | 3 | 179 | chitobiosyldiphosphodolichol beta-mannosyltransferase and similar proteins. This family is most closely related to the GT33 family of glycosyltransferases. The yeast gene ALG1 has been shown to function as a mannosyltransferase that catalyzes the formation of dolichol pyrophosphate (Dol-PP)-GlcNAc2Man from GDP-Man and Dol-PP-Glc-NAc2, and participates in the formation of the lipid-linked precursor oligosaccharide for N-glycosylation. In humans ALG1 has been associated with the congenital disorders of glycosylation (CDG) designated as subtype CDG-Ik. |
| 215155 | PLN02275 | 3.61e-50 | 37 | 193 | 4 | 160 | transferase, transferring glycosyl groups |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.90e-70 | 12 | 193 | 5 | 186 | |
| 2.75e-44 | 25 | 194 | 30 | 199 | |
| 2.75e-44 | 25 | 194 | 30 | 199 | |
| 2.75e-44 | 25 | 194 | 30 | 199 | |
| 2.75e-44 | 25 | 194 | 30 | 199 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.27e-31 | 29 | 196 | 16 | 183 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg1 PE=3 SV=2 |
|
| 1.44e-30 | 34 | 210 | 37 | 213 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=ALG1 PE=3 SV=1 |
|
| 5.29e-30 | 38 | 210 | 33 | 200 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Pongo abelii OX=9601 GN=ALG1 PE=2 SV=1 |
|
| 6.98e-29 | 40 | 192 | 7 | 159 | UDP-glycosyltransferase TURAN OS=Arabidopsis thaliana OX=3702 GN=TUN PE=2 SV=1 |
|
| 9.52e-29 | 38 | 210 | 33 | 200 | Chitobiosyldiphosphodolichol beta-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
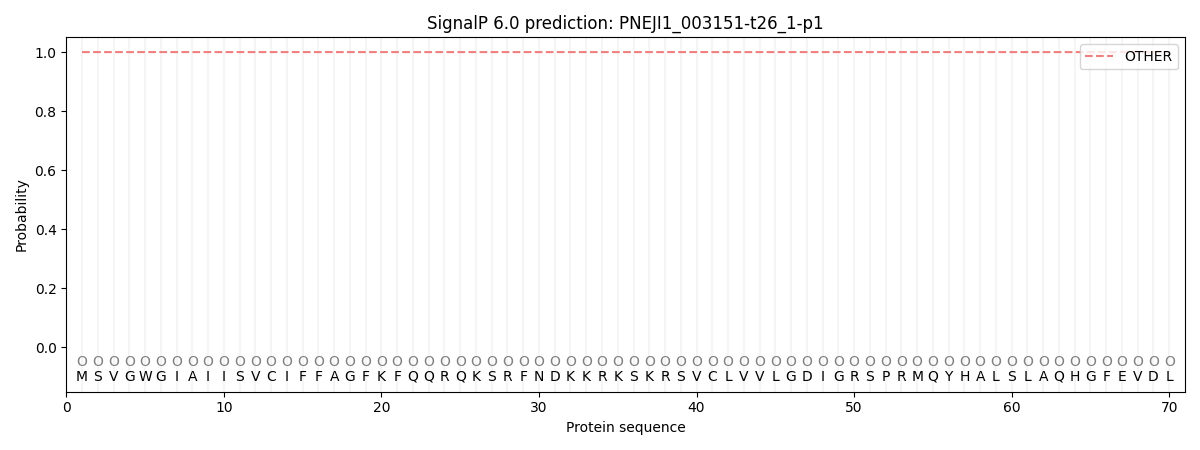
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000048 | 0.000001 |