You are browsing environment: FUNGIDB
CAZyme Information: PNEJI1_002552-t26_1-p1
You are here: Home > Sequence: PNEJI1_002552-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
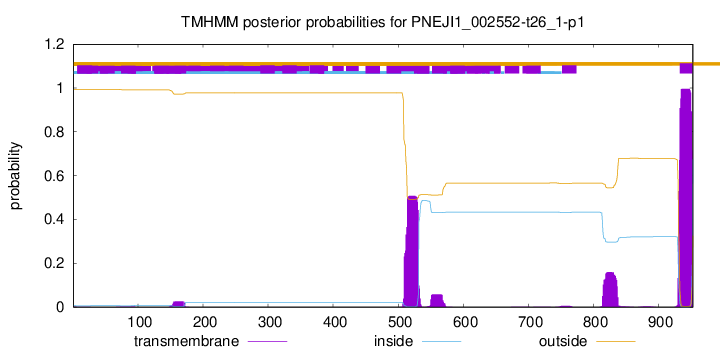
TMHMM annotations
Basic Information help
| Species | Pneumocystis jirovecii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis jirovecii | |||||||||||
| CAZyme ID | PNEJI1_002552-t26_1-p1 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 537 | 892 | 1.8e-91 | 0.946927374301676 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215241 | PLN02440 | 0.0 | 1 | 477 | 1 | 464 | amidophosphoribosyltransferase |
| 273461 | purF | 0.0 | 2 | 464 | 1 | 442 | amidophosphoribosyltransferase. Alternate name: glutamine phosphoribosylpyrophosphate (PRPP) amidotransferase. [Purines, pyrimidines, nucleosides, and nucleotides, Purine ribonucleotide biosynthesis] |
| 223112 | PurF | 0.0 | 1 | 479 | 4 | 468 | Glutamine phosphoribosylpyrophosphate amidotransferase [Nucleotide transport and metabolism]. |
| 236427 | PRK09246 | 0.0 | 1 | 499 | 1 | 501 | amidophosphoribosyltransferase; Provisional |
| 397638 | Glyco_hydro_76 | 1.15e-153 | 538 | 880 | 1 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 19 | 953 | 7 | 946 | |
| 9.90e-160 | 1 | 476 | 1 | 480 | |
| 3.19e-126 | 531 | 953 | 23 | 450 | |
| 4.48e-126 | 531 | 953 | 23 | 450 | |
| 9.62e-126 | 531 | 952 | 34 | 461 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.05e-153 | 2 | 478 | 1 | 481 | The structure of E. coli PurF in complex with ppGpp-Mg [Escherichia coli K-12],6CZF_B The structure of E. coli PurF in complex with ppGpp-Mg [Escherichia coli K-12],6CZF_C The structure of E. coli PurF in complex with ppGpp-Mg [Escherichia coli K-12],6CZF_D The structure of E. coli PurF in complex with ppGpp-Mg [Escherichia coli K-12] |
|
| 1.28e-153 | 2 | 478 | 1 | 481 | Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Gmp, 1 Mg Per Subunit [Escherichia coli],1ECB_B Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Gmp, 1 Mg Per Subunit [Escherichia coli],1ECB_C Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Gmp, 1 Mg Per Subunit [Escherichia coli],1ECB_D Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Gmp, 1 Mg Per Subunit [Escherichia coli],1ECC_A Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With Mn-Cprpp And 5-Oxo- Norleucine [Escherichia coli],1ECC_B Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With Mn-Cprpp And 5-Oxo- Norleucine [Escherichia coli],1ECF_A Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase [Escherichia coli],1ECF_B Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase [Escherichia coli],1ECG_A DON INACTIVATED ESCHERICHIA COLI GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE (PRPP) AMIDOTRANSFERASE [Escherichia coli],1ECG_B DON INACTIVATED ESCHERICHIA COLI GLUTAMINE PHOSPHORIBOSYLPYROPHOSPHATE (PRPP) AMIDOTRANSFERASE [Escherichia coli],1ECJ_A Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Amp Per Tetramer [Escherichia coli],1ECJ_B Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Amp Per Tetramer [Escherichia coli],1ECJ_C Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Amp Per Tetramer [Escherichia coli],1ECJ_D Escherichia Coli Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase Complexed With 2 Amp Per Tetramer [Escherichia coli],6OTT_A Structure of PurF in complex with ppApp [Escherichia coli K-12],6OTT_B Structure of PurF in complex with ppApp [Escherichia coli K-12] |
|
| 2.83e-94 | 530 | 908 | 27 | 423 | Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY1_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY2_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY5_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY6_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY7_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495] |
|
| 6.32e-68 | 2 | 467 | 1 | 449 | Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase From B. Subtilis Complexed With Adp And Gmp [Bacillus subtilis],1AO0_B Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase From B. Subtilis Complexed With Adp And Gmp [Bacillus subtilis],1AO0_C Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase From B. Subtilis Complexed With Adp And Gmp [Bacillus subtilis],1AO0_D Glutamine Phosphoribosylpyrophosphate (Prpp) Amidotransferase From B. Subtilis Complexed With Adp And Gmp [Bacillus subtilis] |
|
| 7.37e-68 | 2 | 467 | 1 | 449 | STRUCTURE OF THE ALLOSTERIC REGULATORY ENZYME OF PURINE BIOSYNTHESIS [Bacillus subtilis],1GPH_2 STRUCTURE OF THE ALLOSTERIC REGULATORY ENZYME OF PURINE BIOSYNTHESIS [Bacillus subtilis],1GPH_3 STRUCTURE OF THE ALLOSTERIC REGULATORY ENZYME OF PURINE BIOSYNTHESIS [Bacillus subtilis],1GPH_4 STRUCTURE OF THE ALLOSTERIC REGULATORY ENZYME OF PURINE BIOSYNTHESIS [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.80e-201 | 1 | 511 | 1 | 523 | Amidophosphoribosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ade4 PE=1 SV=1 |
|
| 9.13e-190 | 1 | 511 | 1 | 506 | Amidophosphoribosyltransferase OS=Lachancea kluyveri OX=4934 GN=ADE4 PE=3 SV=1 |
|
| 2.30e-187 | 1 | 511 | 1 | 506 | Amidophosphoribosyltransferase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=ADE4 PE=1 SV=2 |
|
| 1.82e-158 | 1 | 477 | 1 | 479 | Amidophosphoribosyltransferase OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=purF PE=3 SV=3 |
|
| 7.97e-158 | 1 | 465 | 1 | 468 | Amidophosphoribosyltransferase OS=Pasteurella multocida (strain Pm70) OX=272843 GN=purF PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
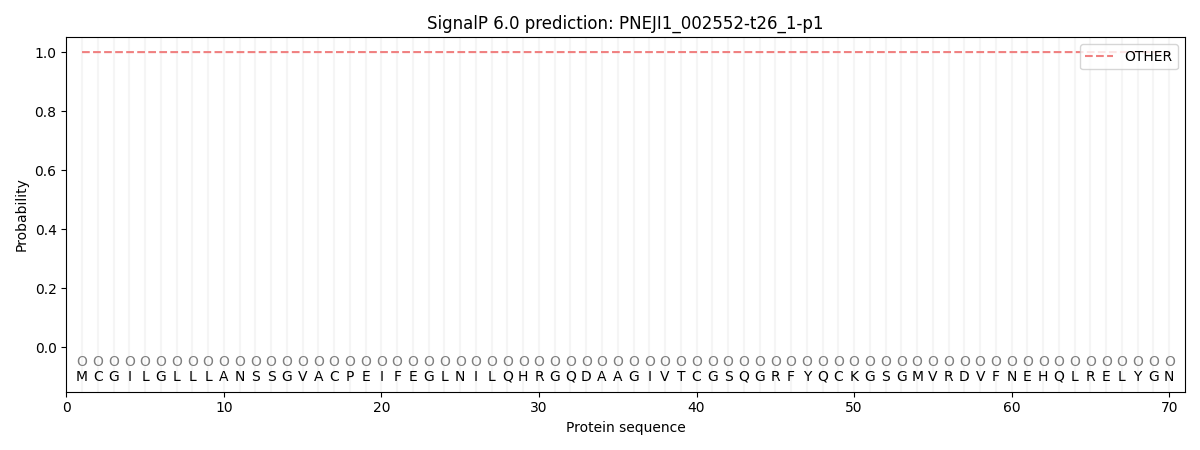
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000005 | 0.000059 |