You are browsing environment: FUNGIDB
CAZyme Information: PNEJI1_001061-t26_1-p1
You are here: Home > Sequence: PNEJI1_001061-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis jirovecii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis jirovecii | |||||||||||
| CAZyme ID | PNEJI1_001061-t26_1-p1 | |||||||||||
| CAZy Family | GH47 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.34:56 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 856 | 1632 | 0 | 0.9837618403247632 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 856 | 1682 | 1 | 819 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 1.06e-45 | 374 | 482 | 1 | 110 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1918 | 35 | 1979 | |
| 0.0 | 1 | 1918 | 1 | 1942 | |
| 0.0 | 101 | 1918 | 96 | 1942 | |
| 0.0 | 48 | 1895 | 6 | 1875 | |
| 0.0 | 47 | 1896 | 3 | 1884 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 57 | 1903 | 21 | 1871 | 1,3-beta-glucan synthase component GSC2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GSC2 PE=1 SV=2 |
|
| 0.0 | 129 | 1875 | 86 | 1850 | 1,3-beta-glucan synthase component FKS1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=fksA PE=3 SV=1 |
|
| 0.0 | 227 | 1903 | 15 | 1764 | 1,3-beta-glucan synthase component FKS3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS3 PE=1 SV=1 |
|
| 0.0 | 52 | 1903 | 10 | 1852 | 1,3-beta-glucan synthase component FKS1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS1 PE=1 SV=2 |
|
| 0.0 | 187 | 1884 | 103 | 1794 | 1,3-beta-glucan synthase component FKS1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=FKS1 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000055 | 0.000003 |
TMHMM Annotations download full data without filtering help
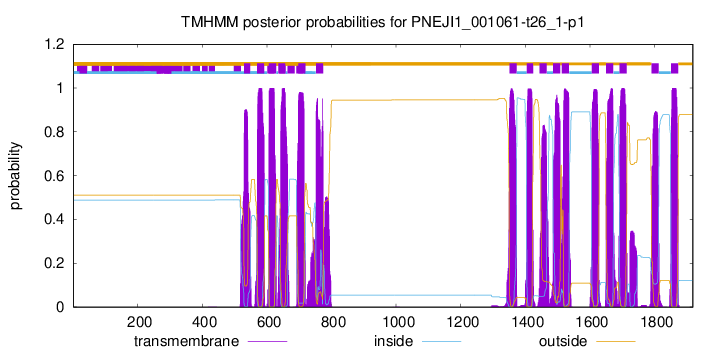
| Start | End |
|---|---|
| 529 | 548 |
| 569 | 591 |
| 606 | 628 |
| 635 | 657 |
| 697 | 719 |
| 752 | 774 |
| 1351 | 1373 |
| 1405 | 1424 |
| 1444 | 1466 |
| 1486 | 1508 |
| 1513 | 1535 |
| 1606 | 1628 |
| 1650 | 1672 |
| 1692 | 1714 |
| 1791 | 1813 |
| 1851 | 1873 |
