You are browsing environment: FUNGIDB
PMAA_101960-t26_1-p1
Basic Information
help
Species
Talaromyces marneffei
Lineage
Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces marneffei
CAZyme ID
PMAA_101960-t26_1-p1
CAZy Family
GT4
CAZyme Description
conserved hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
244
DS995902|CGC15 26495.49
5.9053
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_TmarneffeiATCC18224
10194
441960
145
10049
Gene Location
No EC number prediction in PMAA_101960-t26_1-p1.
Family
Start
End
Evalue
family coverage
PL7
31
243
2.7e-57
0.985981308411215
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
400923
Alginate_lyase2
7.15e-37
31
243
3
222
Alginate lyase. Alginate lyases are enzymes that degrade the linear polysaccharide alignate. They cleave the glycosidic linkage of alignate through a beta-elimination reaction. This family forms an all beta fold and is different to all alpha fold of pfam05426.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.30e-180
1
244
1
244
4.28e-96
19
243
29
252
1.86e-63
11
243
12
248
3.60e-62
21
243
24
254
9.98e-61
11
243
12
249
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
2.05e-59
21
243
6
231
Chain A, Glucuronan lyase [Trichoderma parareesei]
more
PMAA_101960-t26_1-p1 has no Swissprot hit.
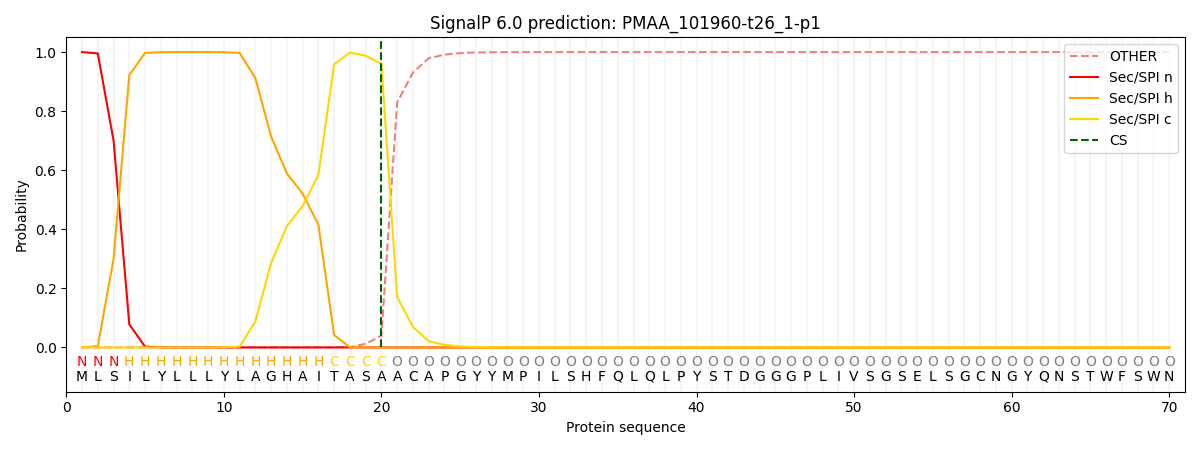
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000214
0.999780
CS pos: 20-21. Pr: 0.9601