You are browsing environment: FUNGIDB
CAZyme Information: PMAA_101130-t26_1-p1
You are here: Home > Sequence: PMAA_101130-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
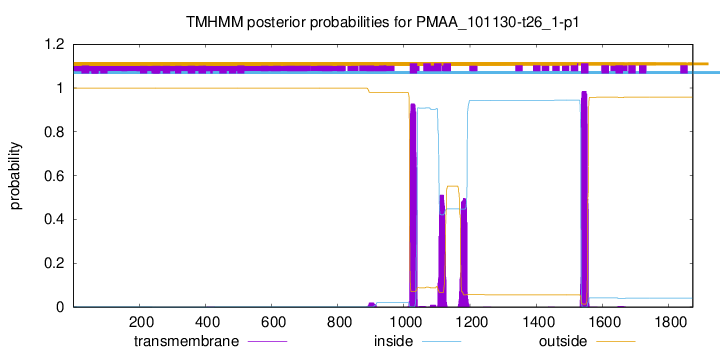
TMHMM annotations
Basic Information help
| Species | Talaromyces marneffei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces marneffei | |||||||||||
| CAZyme ID | PMAA_101130-t26_1-p1 | |||||||||||
| CAZy Family | GT34 | |||||||||||
| CAZyme Description | alpha-1,6-mannosyltransferase subunit, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 1568 | 1828 | 1.9e-82 | 0.983739837398374 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 350766 | DEXDc_SHPRH-like | 1.59e-83 | 348 | 620 | 1 | 241 | DEXH-box helicase domain of SHPRH-like proteins. The SHPRH-like subgroup belongs to the DEAD-like helicase superfamily, a diverse family of proteins involved in ATP-dependent RNA or DNA unwinding. This domain contains the ATP-binding region. |
| 368535 | Glyco_transf_34 | 3.39e-79 | 1568 | 1826 | 4 | 238 | galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34. |
| 223627 | HepA | 4.34e-61 | 395 | 977 | 350 | 847 | Superfamily II DNA or RNA helicase, SNF2 family [Transcription, Replication, recombination and repair]. |
| 410683 | CYP57A1-like | 1.45e-59 | 1078 | 1283 | 108 | 355 | cytochrome P450 family 57, subfamily A, polypeptide 1 and similar cytochrome P450s. This family is composed of fungal cytochrome P450s including: Nectria haematococca cytochrome P450 57A1 (CYP57A1), also called pisatin demethylase, which detoxifies the phytoalexin pisatin; Penicillium aethiopicum P450 monooxygenase gsfF, also called griseofulvin synthesis protein F, which catalyzes the coupling of orcinol and phloroglucinol rings in griseophenone B to form desmethyl-dehydrogriseofulvin A during the biosynthesis of griseofulvin, a spirocyclic fungal natural product used to treat dermatophyte infections; and Penicillium aethiopicum P450 monooxygenase vrtE, also called viridicatumtoxin synthesis protein E, which catalyzes hydroxylation at C5 of the polyketide backbone during the biosynthesis of viridicatumtoxin, a tetracycline-like fungal meroterpenoid. The CYP57A1-like family belongs to the large cytochrome P450 (P450, CYP) superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. |
| 350829 | DEXHc_HLTF1_SMARC3 | 8.69e-55 | 351 | 620 | 4 | 239 | DEXH-box helicase domain of HLTF1. Helicase like transcription factor (HLTF1, also known as HIP116 or SMARCA3) has both helicase and E3 ubiquitin ligase activities and ATP-dependent nucleosome-remodeling activity. HLTF1 is a member of the DEAD-like helicase superfamily, a diverse family of proteins involved in ATP-dependent RNA or DNA unwinding. This domain contains the ATP-binding region. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1164 | 1874 | 1 | 603 | |
| 1.74e-223 | 1339 | 1873 | 285 | 812 | |
| 2.07e-186 | 1407 | 1861 | 1 | 444 | |
| 3.84e-186 | 1407 | 1861 | 1 | 452 | |
| 1.60e-181 | 1407 | 1854 | 1 | 439 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.35e-56 | 347 | 965 | 233 | 925 | Chain A, DNA repair protein RAD5 [Kluyveromyces lactis NRRL Y-1140],6L8O_A Chain A, DNA repair protein RAD5 [Kluyveromyces lactis NRRL Y-1140] |
|
| 1.81e-35 | 401 | 978 | 1313 | 1804 | Chain B, Helicase-like protein [Thermochaetoides thermophila DSM 1495] |
|
| 1.55e-29 | 406 | 965 | 127 | 551 | Crystal structure of MtISWI [Thermothelomyces thermophilus ATCC 42464],5JXR_B Crystal structure of MtISWI [Thermothelomyces thermophilus ATCC 42464] |
|
| 3.00e-29 | 406 | 965 | 124 | 548 | Chain K, chromatin remodeling factor ISWI [Thermochaetoides thermophila] |
|
| 1.30e-28 | 406 | 965 | 36 | 457 | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h bound at SHL-2 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.08e-91 | 1559 | 1847 | 112 | 389 | Probable alpha-1,6-mannosyltransferase MNN10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN10 PE=1 SV=1 |
|
| 9.73e-79 | 1569 | 1848 | 75 | 343 | Uncharacterized alpha-1,2-galactosyltransferase C637.06 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC637.06 PE=2 SV=2 |
|
| 1.77e-71 | 338 | 973 | 219 | 859 | Putative SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A member 3-like 1 OS=Arabidopsis thaliana OX=3702 GN=At5g05130 PE=2 SV=1 |
|
| 6.92e-70 | 335 | 990 | 603 | 1273 | DNA repair protein RAD5B OS=Arabidopsis thaliana OX=3702 GN=RAD5B PE=3 SV=1 |
|
| 9.27e-67 | 335 | 990 | 335 | 1025 | DNA repair protein RAD5A OS=Arabidopsis thaliana OX=3702 GN=RAD5A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000044 | 0.000000 |