You are browsing environment: FUNGIDB
CAZyme Information: PMAA_064660-t26_1-p1
You are here: Home > Sequence: PMAA_064660-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Talaromyces marneffei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces marneffei | |||||||||||
| CAZyme ID | PMAA_064660-t26_1-p1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | beta-mannosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.25:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 20 | 774 | 9.1e-74 | 0.6715425531914894 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225789 | LacZ | 2.70e-43 | 96 | 733 | 83 | 636 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| 407625 | Ig_mannosidase | 2.97e-16 | 847 | 924 | 1 | 78 | Ig-fold domain. This Ig-like fold domain is found in mannosidase enzymes. |
| 236673 | ebgA | 2.03e-10 | 206 | 477 | 202 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| 395572 | Glyco_hydro_2 | 3.02e-08 | 217 | 337 | 6 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| 407658 | Mannosidase_ig | 1.16e-07 | 740 | 814 | 1 | 71 | Mannosidase Ig/CBM-like domain. This domain corresponds to domain 4 in the structure of Bacteroides thetaiotaomicron beta-mannosidase, BtMan2A. This domain has an Ig-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 953 | 1 | 958 | |
| 0.0 | 6 | 926 | 5 | 922 | |
| 0.0 | 8 | 941 | 10 | 943 | |
| 0.0 | 8 | 941 | 10 | 943 | |
| 0.0 | 8 | 925 | 10 | 910 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 24 | 924 | 32 | 938 | Structure of Fungal beta-mannosidase from Glycoside Hydrolase Family 2 of Trichoderma harzianum [Trichoderma harzianum],4UOJ_A Chain A, BETA-MANNOSIDASE GH2 [Trichoderma harzianum],4UOJ_B Chain B, BETA-MANNOSIDASE GH2 [Trichoderma harzianum] |
|
| 3.20e-98 | 25 | 893 | 14 | 836 | mouse beta-mannosidase (MANBA) [Mus musculus],6DDU_A mouse beta-mannosidase bound to beta-D-mannose (MANBA) [Mus musculus] |
|
| 1.26e-73 | 42 | 905 | 34 | 835 | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
|
| 1.28e-73 | 42 | 905 | 34 | 835 | Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
|
| 8.02e-73 | 42 | 905 | 32 | 833 | Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 0.0 | 14 | 925 | 16 | 922 | Beta-mannosidase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mndA PE=3 SV=1 |
|
| 0.0 | 13 | 928 | 17 | 936 | Beta-mannosidase A OS=Aspergillus aculeatus OX=5053 GN=mndA PE=1 SV=1 |
|
| 0.0 | 21 | 925 | 24 | 928 | Beta-mannosidase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=mndA PE=3 SV=2 |
|
| 0.0 | 8 | 925 | 10 | 910 | Beta-mannosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=mndA PE=3 SV=1 |
|
| 0.0 | 8 | 925 | 11 | 928 | Beta-mannosidase A OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=mndA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
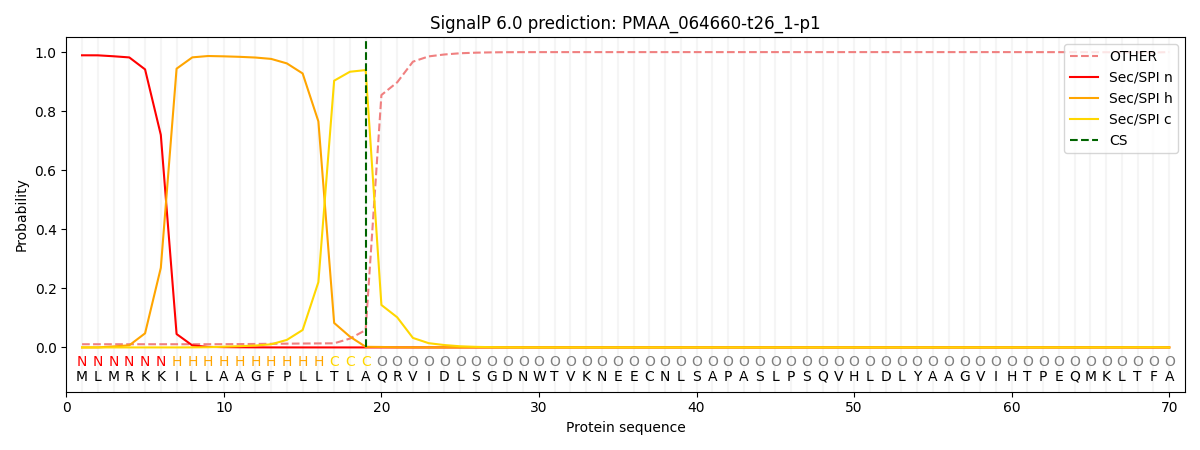
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.015751 | 0.984235 | CS pos: 19-20. Pr: 0.9391 |
