You are browsing environment: FUNGIDB
CAZyme Information: PMAA_050670-t26_1-p1
You are here: Home > Sequence: PMAA_050670-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Talaromyces marneffei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces marneffei | |||||||||||
| CAZyme ID | PMAA_050670-t26_1-p1 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | rhamnogalacturonase A precursor, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.171:11 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 72 | 375 | 9.5e-47 | 0.9446153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215540 | PLN03010 | 6.30e-16 | 44 | 377 | 48 | 391 | polygalacturonase |
| 178580 | PLN03003 | 3.63e-15 | 44 | 377 | 25 | 377 | Probable polygalacturonase At3g15720 |
| 395231 | Glyco_hydro_28 | 6.76e-14 | 143 | 375 | 79 | 315 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| 215120 | PLN02188 | 2.71e-12 | 36 | 375 | 30 | 390 | polygalacturonase/glycoside hydrolase family protein |
| 177865 | PLN02218 | 6.93e-11 | 45 | 296 | 70 | 344 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 447 | 1 | 447 | |
| 1.01e-275 | 1 | 445 | 1 | 437 | |
| 1.77e-215 | 2 | 445 | 4 | 446 | |
| 1.29e-210 | 1 | 445 | 3 | 446 | |
| 1.29e-210 | 1 | 445 | 3 | 446 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.10e-194 | 28 | 439 | 6 | 416 | Rhamnogalacturonase A From Aspergillus Aculeatus [Aspergillus aculeatus] |
|
| 2.87e-14 | 39 | 374 | 15 | 373 | Crystal structure of endo-xylogalacturonan hydrolase from Aspergillus tubingensis [Aspergillus tubingensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.34e-194 | 11 | 439 | 11 | 434 | Rhamnogalacturonase A OS=Aspergillus aculeatus OX=5053 GN=rhgA PE=1 SV=1 |
|
| 1.57e-187 | 28 | 439 | 27 | 439 | Probable rhamnogalacturonase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=rhgB PE=3 SV=1 |
|
| 3.05e-186 | 28 | 439 | 27 | 439 | Probable rhamnogalacturonase B OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=rhgB PE=3 SV=1 |
|
| 6.13e-186 | 28 | 439 | 27 | 439 | Probable rhamnogalacturonase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=rhgB PE=3 SV=1 |
|
| 2.93e-184 | 14 | 439 | 17 | 441 | Probable rhamnogalacturonase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=rhgB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
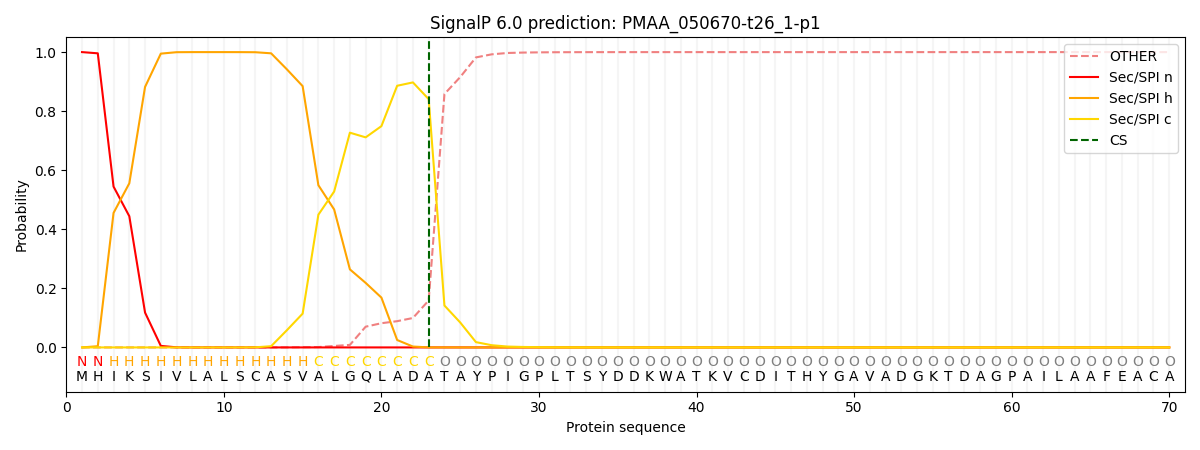
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000246 | 0.999721 | CS pos: 23-24. Pr: 0.8408 |
