You are browsing environment: FUNGIDB
CAZyme Information: PMAA_038960-t26_1-p1
You are here: Home > Sequence: PMAA_038960-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
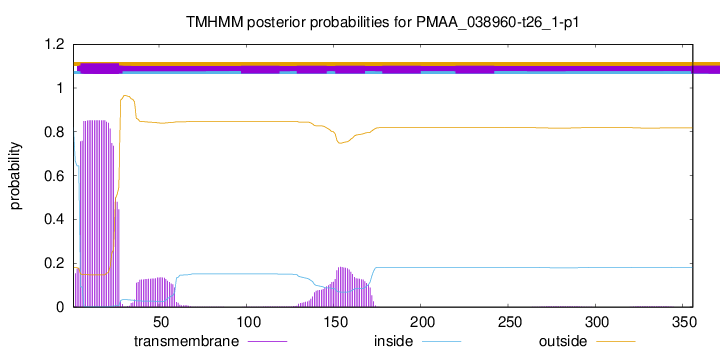
TMHMM annotations
Basic Information help
| Species | Talaromyces marneffei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Trichocomaceae; Talaromyces; Talaromyces marneffei | |||||||||||
| CAZyme ID | PMAA_038960-t26_1-p1 | |||||||||||
| CAZy Family | GH15 | |||||||||||
| CAZyme Description | acetyl xylan esterase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.72:3 | 1.14.99.56:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 34 | 188 | 4.3e-19 | 0.5947136563876652 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 273828 | esterase_phb | 2.81e-38 | 36 | 250 | 1 | 209 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| 226040 | LpqC | 2.77e-27 | 1 | 279 | 14 | 277 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| 402228 | Esterase_phd | 1.37e-16 | 38 | 241 | 5 | 201 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| 395595 | CBM_1 | 5.36e-13 | 324 | 352 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 5.47e-13 | 323 | 356 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.81e-247 | 1 | 356 | 1 | 356 | |
| 1.22e-230 | 1 | 356 | 1 | 357 | |
| 1.22e-230 | 1 | 356 | 1 | 357 | |
| 2.12e-230 | 1 | 356 | 1 | 353 | |
| 6.51e-224 | 1 | 356 | 1 | 349 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.34e-78 | 18 | 290 | 1 | 273 | Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
|
| 7.72e-14 | 320 | 356 | 2 | 38 | Chain A, ENDOGLUCANASE EG-1 [Trichoderma reesei] |
|
| 7.82e-11 | 322 | 356 | 2 | 36 | Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 2.75e-10 | 323 | 356 | 3 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
|
| 1.03e-09 | 322 | 356 | 372 | 406 | GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8M_B GH10 endo-xylanase [Aspergillus aculeatus ATCC 16872],6Q8N_A GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872],6Q8N_B GH10 endo-xylanase in complex with xylobiose epoxide inhibitor [Aspergillus aculeatus ATCC 16872] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.77e-231 | 1 | 356 | 1 | 353 | Feruloyl esterase B OS=Talaromyces funiculosus OX=28572 GN=FAEB PE=1 SV=1 |
|
| 2.47e-212 | 1 | 356 | 1 | 358 | Feruloyl esterase B OS=Talaromyces stipitatus (strain ATCC 10500 / CBS 375.48 / QM 6759 / NRRL 1006) OX=441959 GN=faeB PE=1 SV=1 |
|
| 1.50e-94 | 17 | 291 | 17 | 290 | Feruloyl esterase B OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=fae-1 PE=2 SV=2 |
|
| 3.02e-92 | 16 | 356 | 29 | 371 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
|
| 2.20e-91 | 16 | 356 | 29 | 368 | Probable acetylxylan esterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=axeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000261 | 0.999745 | CS pos: 18-19. Pr: 0.9769 |