You are browsing environment: FUNGIDB
CAZyme Information: PIW_T012305-RA-p1
You are here: Home > Sequence: PIW_T012305-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium iwayamae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium iwayamae | |||||||||||
| CAZyme ID | PIW_T012305-RA-p1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | Exo-1,3-beta-glucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 379 | 657 | 2.7e-44 | 0.9711191335740073 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396255 | Amino_oxidase | 1.94e-47 | 89 | 330 | 196 | 435 | Flavin containing amine oxidoreductase. This family consists of various amine oxidases, including maze polyamine oxidase (PAO) and various flavin containing monoamine oxidases (MAO). The aligned region includes the flavin binding site of these enzymes. The family also contains phytoene dehydrogenases and related enzymes. In vertebrates MAO plays an important role regulating the intracellular levels of amines via there oxidation; these include various neurotransmitters, neurotoxins and trace amines. In lower eukaryotes such as aspergillus and in bacteria the main role of amine oxidases is to provide a source of ammonium. PAOs in plants, bacteria and protozoa oxidase spermidine and spermine to an aminobutyral, diaminopropane and hydrogen peroxide and are involved in the catabolism of polyamines. Other members of this family include tryptophan 2-monooxygenase, putrescine oxidase, corticosteroid binding proteins and antibacterial glycoproteins. |
| 177909 | PLN02268 | 3.49e-39 | 55 | 347 | 160 | 434 | probable polyamine oxidase |
| 215187 | PLN02328 | 1.58e-38 | 86 | 340 | 420 | 674 | lysine-specific histone demethylase 1 homolog |
| 178578 | PLN03000 | 4.34e-34 | 86 | 340 | 364 | 618 | amine oxidase |
| 215527 | PLN02976 | 1.08e-30 | 82 | 318 | 919 | 1160 | amine oxidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 8.86e-179 | 339 | 677 | 5 | 369 | |
| 1.64e-177 | 353 | 677 | 37 | 361 | |
| 4.20e-125 | 351 | 676 | 46 | 372 | |
| 2.01e-108 | 351 | 676 | 136 | 452 | |
| 2.56e-94 | 353 | 677 | 66 | 387 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.20e-24 | 55 | 318 | 490 | 751 | Crystal structure of LSD2 with H3 [Homo sapiens],4GU0_B Crystal structure of LSD2 with H3 [Homo sapiens],4GU0_C Crystal structure of LSD2 with H3 [Homo sapiens],4GU0_D Crystal structure of LSD2 with H3 [Homo sapiens],4GUR_A Crystal structure of LSD2-NPAC with H3 in space group P21 [Homo sapiens],4GUS_A Crystal structure of LSD2-NPAC with H3 in space group P3221 [Homo sapiens],4GUT_A Crystal structure of LSD2-NPAC [Homo sapiens],4GUU_A Crystal structure of LSD2-NPAC with tranylcypromine [Homo sapiens],4HSU_A Crystal structure of LSD2-NPAC with H3(1-26)in space group P21 [Homo sapiens],6R1U_K Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 [Homo sapiens],6R25_K Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 3 [Homo sapiens] |
|
| 5.27e-24 | 55 | 318 | 498 | 759 | Crystal structure of LSD2 [Homo sapiens],4GU1_B Crystal structure of LSD2 [Homo sapiens] |
|
| 5.37e-24 | 55 | 318 | 510 | 771 | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A [Homo sapiens],4FWF_A Complex structure of LSD2/AOF1/KDM1b with H3K4 mimic [Homo sapiens],4FWJ_A Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A [Homo sapiens],4FWJ_B Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A [Homo sapiens] |
|
| 6.11e-21 | 89 | 330 | 391 | 644 | Chain A, Lysine-specific histone demethylase 1 [Homo sapiens] |
|
| 1.85e-20 | 89 | 330 | 390 | 643 | Crystal structure of Human LSD1 [Homo sapiens],6NQU_A Human LSD1 in complex with GSK2879552 [Homo sapiens],6NR5_A Human LSD1 in complex with Phenelzine sulfate [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.32e-31 | 27 | 340 | 399 | 702 | Lysine-specific histone demethylase 1 homolog 1 OS=Arabidopsis thaliana OX=3702 GN=LDL1 PE=1 SV=1 |
|
| 5.62e-30 | 39 | 355 | 414 | 725 | Lysine-specific histone demethylase 1 homolog 1 OS=Oryza sativa subsp. japonica OX=39947 GN=Os02g0755200 PE=2 SV=1 |
|
| 8.94e-29 | 86 | 352 | 377 | 644 | Lysine-specific histone demethylase 1 homolog 3 OS=Oryza sativa subsp. indica OX=39946 GN=B0103C08-B0602B01.13 PE=3 SV=1 |
|
| 1.19e-28 | 82 | 352 | 381 | 644 | Lysine-specific histone demethylase 1 homolog 3 OS=Oryza sativa subsp. japonica OX=39947 GN=Os04g0560300 PE=2 SV=2 |
|
| 3.17e-27 | 55 | 357 | 183 | 467 | Polyamine oxidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=PAO4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
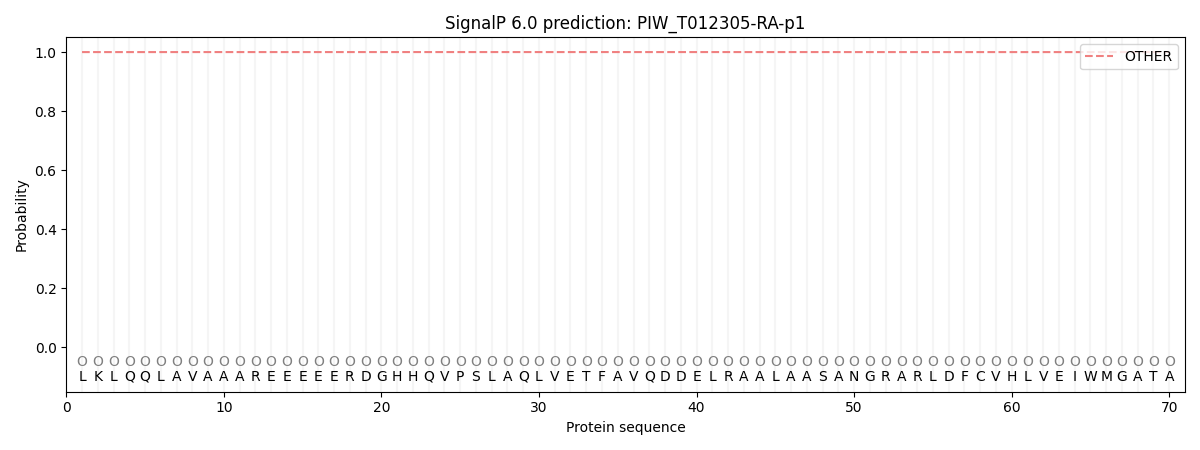
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000080 | 0.000002 |
