You are browsing environment: FUNGIDB
CAZyme Information: PIW_T007930-RA-p1
You are here: Home > Sequence: PIW_T007930-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium iwayamae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium iwayamae | |||||||||||
| CAZyme ID | PIW_T007930-RA-p1 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | Beta-glucosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:2 | 3.2.1.23:1 | 3.2.1.38:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 26 | 280 | 4.6e-87 | 0.5594405594405595 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 1.45e-95 | 29 | 280 | 5 | 244 | Glycosyl hydrolase family 1. |
| 274539 | BGL | 1.38e-92 | 30 | 276 | 1 | 235 | beta-galactosidase. |
| 225343 | BglB | 4.14e-88 | 29 | 280 | 4 | 247 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215435 | PLN02814 | 9.50e-62 | 1 | 283 | 1 | 281 | beta-glucosidase |
| 184102 | PRK13511 | 1.80e-59 | 29 | 277 | 5 | 238 | 6-phospho-beta-galactosidase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.36e-130 | 6 | 283 | 4 | 286 | |
| 2.87e-125 | 28 | 283 | 12 | 267 | |
| 2.69e-108 | 2 | 283 | 3 | 277 | |
| 1.15e-71 | 77 | 283 | 5 | 214 | |
| 2.32e-64 | 26 | 283 | 28 | 292 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.25e-64 | 29 | 280 | 11 | 251 | Chain A, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1H_B Chain B, ANCESTRAL RECONSTRUCTED GLYCOSIDASE [synthetic construct],6Z1M_A Chain A, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_B Chain B, Ancestral reconstructed glycosidase [synthetic construct],6Z1M_C Chain C, Ancestral reconstructed glycosidase [synthetic construct] |
|
| 4.38e-61 | 19 | 280 | 5 | 253 | Chain AAA, Beta-glucosidase [Alicyclobacillus tengchongensis],6ZIV_BBB Chain BBB, Beta-glucosidase [Alicyclobacillus tengchongensis],6ZIV_CCC Chain CCC, Beta-glucosidase [Alicyclobacillus tengchongensis],6ZIV_DDD Chain DDD, Beta-glucosidase [Alicyclobacillus tengchongensis],6ZIV_EEE Chain EEE, Beta-glucosidase [Alicyclobacillus tengchongensis],6ZIV_FFF Chain FFF, Beta-glucosidase [Alicyclobacillus tengchongensis],6ZIV_GGG Chain GGG, Beta-glucosidase [Alicyclobacillus tengchongensis],6ZIV_HHH Chain HHH, Beta-glucosidase [Alicyclobacillus tengchongensis] |
|
| 1.97e-60 | 26 | 283 | 40 | 299 | Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF6_B Structure of inactive mutant of Strictosidine Glucosidase in complex with strictosidine [Rauvolfia serpentina],2JF7_A Structure of Strictosidine Glucosidase [Rauvolfia serpentina],2JF7_B Structure of Strictosidine Glucosidase [Rauvolfia serpentina],3ZJ7_A Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ7_B Crystal structure of strictosidine glucosidase in complex with inhibitor-1 [Rauvolfia serpentina],3ZJ8_A Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina],3ZJ8_B Crystal structure of strictosidine glucosidase in complex with inhibitor-2 [Rauvolfia serpentina] |
|
| 1.52e-59 | 26 | 276 | 3 | 241 | Crystal structure of beta-glucosidase A from bacterium Clostridium cellulovorans [Clostridium cellulovorans],3AHX_B Crystal structure of beta-glucosidase A from bacterium Clostridium cellulovorans [Clostridium cellulovorans],3AHX_C Crystal structure of beta-glucosidase A from bacterium Clostridium cellulovorans [Clostridium cellulovorans],3AHX_D Crystal structure of beta-glucosidase A from bacterium Clostridium cellulovorans [Clostridium cellulovorans] |
|
| 1.68e-58 | 11 | 278 | 15 | 272 | Chain A, Glycoside hydrolase [Nannochloris],5YJ7_B Chain B, Glycoside hydrolase [Nannochloris],5YJ7_C Chain C, Glycoside hydrolase [Nannochloris],5YJ7_D Chain D, Glycoside hydrolase [Nannochloris] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.15e-61 | 26 | 283 | 28 | 292 | Beta-glucosidase 30 OS=Arabidopsis thaliana OX=3702 GN=BGLU30 PE=2 SV=1 |
|
| 6.13e-60 | 25 | 283 | 36 | 300 | Beta-glucosidase 23 OS=Arabidopsis thaliana OX=3702 GN=BGLU23 PE=1 SV=1 |
|
| 1.01e-59 | 26 | 283 | 40 | 299 | Strictosidine-O-beta-D-glucosidase OS=Rauvolfia serpentina OX=4060 GN=SGR1 PE=1 SV=1 |
|
| 1.12e-58 | 26 | 283 | 34 | 298 | Beta-glucosidase 31 OS=Arabidopsis thaliana OX=3702 GN=BGLU31 PE=2 SV=1 |
|
| 1.12e-58 | 26 | 283 | 34 | 298 | Beta-glucosidase 32 OS=Arabidopsis thaliana OX=3702 GN=BGLU32 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
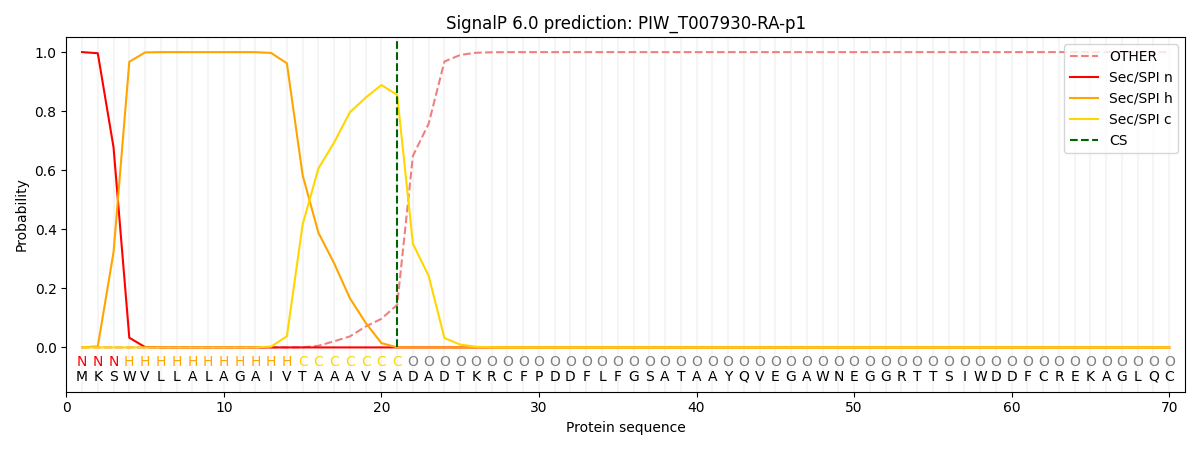
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000223 | 0.999730 | CS pos: 21-22. Pr: 0.8552 |
