You are browsing environment: FUNGIDB
CAZyme Information: PIW_T005133-RA-p1
You are here: Home > Sequence: PIW_T005133-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium iwayamae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium iwayamae | |||||||||||
| CAZyme ID | PIW_T005133-RA-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Carbohydrate-binding protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.99.18:3 | 1.1.99.18:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 18 | 247 | 5.2e-53 | 0.4051094890510949 |
| AA3 | 248 | 506 | 5.9e-44 | 0.4908759124087591 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 1.12e-24 | 26 | 504 | 8 | 524 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 2.05e-14 | 22 | 318 | 2 | 320 | choline dehydrogenase; Validated |
| 238533 | APPLE_Factor_XI_like | 4.33e-13 | 560 | 630 | 2 | 71 | Subfamily of PAN/APPLE-like domains; present in plasma prekallikrein/coagulation factor XI, microneme antigen proteins, and a few prokaryotic proteins. PAN/APPLE domains fulfill diverse biological functions by mediating protein-protein or protein-carbohydrate interactions. |
| 405052 | PAN_4 | 6.68e-08 | 571 | 614 | 2 | 51 | PAN domain. |
| 223566 | TrxB | 1.25e-07 | 23 | 73 | 1 | 51 | Thioredoxin reductase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.72e-216 | 21 | 632 | 24 | 638 | |
| 8.81e-185 | 9 | 631 | 8 | 714 | |
| 9.22e-165 | 9 | 632 | 7 | 719 | |
| 1.09e-145 | 12 | 495 | 4 | 526 | |
| 1.14e-143 | 15 | 503 | 13 | 539 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.03e-40 | 26 | 509 | 3 | 530 | Chain A, Cellobiose dehydrogenase [Phanerodontia chrysosporium],1NAA_B Chain B, Cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 6.44e-40 | 26 | 509 | 8 | 535 | Chain A, cellobiose dehydrogenase [Phanerodontia chrysosporium],1KDG_B Chain B, cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 2.36e-30 | 26 | 503 | 230 | 753 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
|
| 8.25e-27 | 26 | 503 | 8 | 532 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI5_A Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 1.58e-26 | 26 | 503 | 230 | 754 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.25e-38 | 26 | 509 | 235 | 762 | Cellobiose dehydrogenase OS=Phanerodontia chrysosporium OX=2822231 GN=CDH-1 PE=1 SV=1 |
|
| 2.12e-11 | 26 | 491 | 3 | 506 | Oxygen-dependent choline dehydrogenase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=betA PE=3 SV=1 |
|
| 1.12e-10 | 23 | 491 | 2 | 510 | Oxygen-dependent choline dehydrogenase OS=Xanthomonas axonopodis pv. citri (strain 306) OX=190486 GN=betA PE=3 SV=1 |
|
| 2.59e-10 | 26 | 491 | 3 | 506 | Oxygen-dependent choline dehydrogenase OS=Pectobacterium carotovorum subsp. carotovorum (strain PC1) OX=561230 GN=betA PE=3 SV=1 |
|
| 5.97e-10 | 26 | 491 | 7 | 512 | Oxygen-dependent choline dehydrogenase 1 OS=Chromohalobacter salexigens (strain ATCC BAA-138 / DSM 3043 / CIP 106854 / NCIMB 13768 / 1H11) OX=290398 GN=betA1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
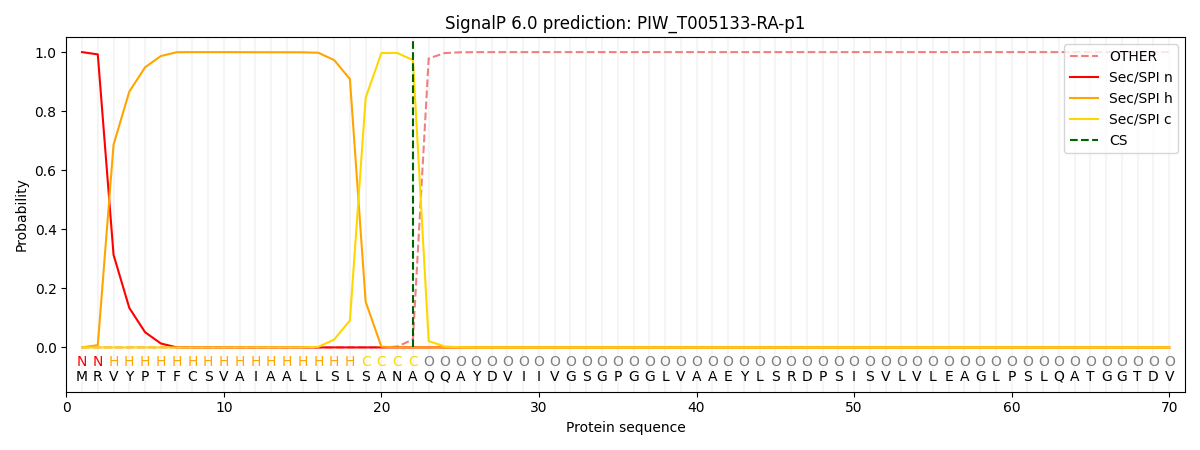
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000574 | 0.999409 | CS pos: 22-23. Pr: 0.9730 |
