You are browsing environment: FUNGIDB
CAZyme Information: PIW_T001244-RA-p1
You are here: Home > Sequence: PIW_T001244-RA-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Globisporangium iwayamae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Pythiaceae; Globisporangium; Globisporangium iwayamae | |||||||||||
| CAZyme ID | PIW_T001244-RA-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | GPI mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT76 | 12 | 439 | 5e-105 | 0.9975429975429976 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 282094 | Mannosyl_trans2 | 4.48e-50 | 11 | 439 | 3 | 439 | Mannosyltransferase (PIG-V). This is a family of eukaryotic ER membrane proteins that are involved in the synthesis of glycosylphosphatidylinositol (GPI), a glycolipid that anchors many proteins to the eukaryotic cell surface. Proteins in this family are involved in transferring the second mannose in the biosynthetic pathway of GPI. |
| 227829 | COG5542 | 1.57e-13 | 55 | 359 | 71 | 349 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.05e-164 | 9 | 439 | 6 | 439 | |
| 8.37e-125 | 9 | 439 | 6 | 490 | |
| 1.57e-55 | 12 | 439 | 15 | 482 | |
| 2.10e-55 | 33 | 439 | 26 | 421 | |
| 6.39e-52 | 12 | 439 | 20 | 497 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.45e-41 | 11 | 439 | 10 | 493 | GPI mannosyltransferase 2 OS=Homo sapiens OX=9606 GN=PIGV PE=1 SV=1 |
|
| 1.91e-40 | 11 | 439 | 10 | 492 | GPI mannosyltransferase 2 OS=Rattus norvegicus OX=10116 GN=Pigv PE=2 SV=1 |
|
| 2.45e-38 | 11 | 439 | 10 | 493 | GPI mannosyltransferase 2 OS=Mus musculus OX=10090 GN=Pigv PE=2 SV=2 |
|
| 1.73e-37 | 52 | 439 | 60 | 452 | GPI mannosyltransferase 2 OS=Drosophila pseudoobscura pseudoobscura OX=46245 GN=GA19757 PE=3 SV=1 |
|
| 7.88e-36 | 52 | 439 | 56 | 449 | GPI mannosyltransferase 2 OS=Drosophila melanogaster OX=7227 GN=PIG-V PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
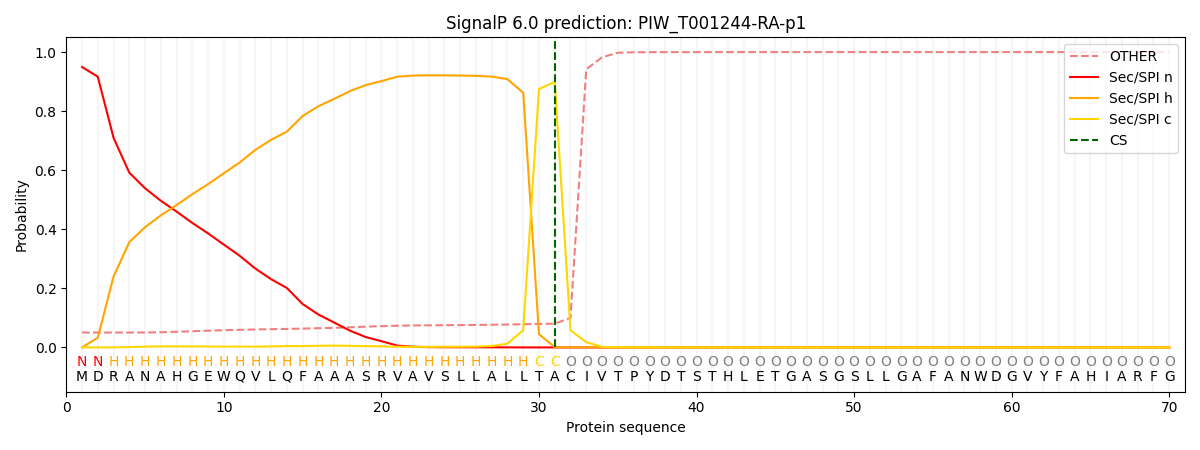
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.050323 | 0.949673 | CS pos: 31-32. Pr: 0.8993 |
TMHMM Annotations download full data without filtering help
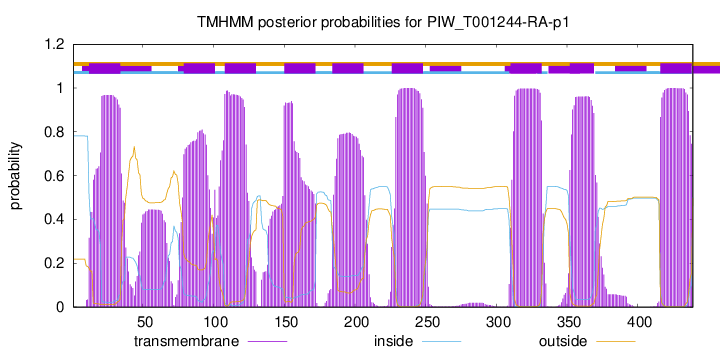
| Start | End |
|---|---|
| 12 | 34 |
| 79 | 101 |
| 108 | 130 |
| 150 | 172 |
| 184 | 206 |
| 226 | 248 |
| 310 | 332 |
| 352 | 369 |
| 416 | 438 |
