You are browsing environment: FUNGIDB
CAZyme Information: PITG_18105-t26_1-p1
You are here: Home > Sequence: PITG_18105-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora infestans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans | |||||||||||
| CAZyme ID | PITG_18105-t26_1-p1 | |||||||||||
| CAZy Family | GT59 | |||||||||||
| CAZyme Description | putative GPI mannosyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 14 | 423 | 3.2e-112 | 0.9897172236503856 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215437 | PLN02816 | 5.57e-117 | 9 | 519 | 40 | 540 | mannosyltransferase |
| 281842 | Glyco_transf_22 | 1.61e-95 | 10 | 423 | 2 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 224720 | ArnT | 0.004 | 94 | 396 | 87 | 390 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| 404170 | PMT_2 | 0.008 | 67 | 212 | 2 | 129 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.75e-169 | 1 | 299 | 1 | 283 | |
| 3.41e-124 | 5 | 525 | 46 | 572 | |
| 1.32e-121 | 12 | 514 | 46 | 531 | |
| 5.98e-120 | 7 | 519 | 40 | 548 | |
| 6.94e-120 | 7 | 519 | 40 | 552 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.80e-119 | 17 | 525 | 47 | 541 | Mannosyltransferase APTG1 OS=Arabidopsis thaliana OX=3702 GN=APTG1 PE=2 SV=1 |
|
| 1.48e-107 | 15 | 528 | 57 | 533 | GPI mannosyltransferase 3 OS=Bos taurus OX=9913 GN=PIGB PE=2 SV=1 |
|
| 1.20e-106 | 17 | 528 | 69 | 543 | GPI mannosyltransferase 3 OS=Homo sapiens OX=9606 GN=PIGB PE=1 SV=1 |
|
| 4.74e-106 | 17 | 507 | 58 | 515 | GPI mannosyltransferase 3 OS=Mus musculus OX=10090 GN=Pigb PE=2 SV=2 |
|
| 1.85e-102 | 13 | 518 | 53 | 523 | GPI mannosyltransferase 3 OS=Xenopus laevis OX=8355 GN=pigb PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999820 | 0.000221 |
TMHMM Annotations download full data without filtering help
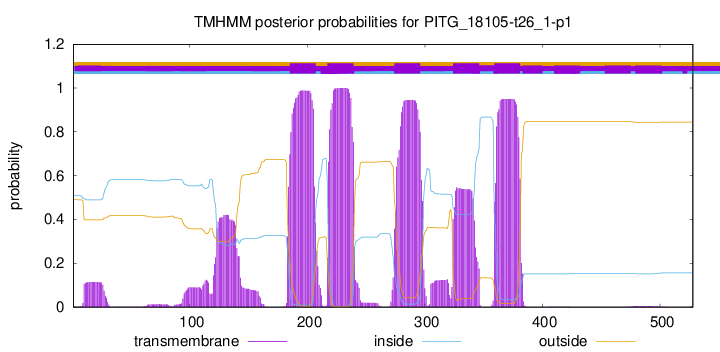
| Start | End |
|---|---|
| 185 | 207 |
| 217 | 239 |
| 274 | 296 |
| 324 | 346 |
| 359 | 381 |
