You are browsing environment: FUNGIDB
CAZyme Information: PITG_11993-t26_1-p1
You are here: Home > Sequence: PITG_11993-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora infestans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans | |||||||||||
| CAZyme ID | PITG_11993-t26_1-p1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | glycoside hydrolase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 36 | 348 | 2.7e-82 | 0.9903225806451613 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 1.45e-31 | 37 | 356 | 42 | 378 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 2.22e-23 | 33 | 341 | 1 | 270 | Cellulase (glycosyl hydrolase family 5). |
| 411343 | exchanger_TraA | 5.62e-05 | 387 | 485 | 305 | 391 | outer membrane exchange protein TraA. TraA, together with its partner TraB, mediates a large scale exchange of outer membrane lipoproteins, and lipids, between closely related strains or clonally identical cells, certain delta-proteobacterial species such as Myxococcus xanthus. The exchange mechanism is likely to involve fusion of outer membrane, probably done to coordinate the social behaviors these bacteria display. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 9 | 501 | 8 | 500 | |
| 9.52e-64 | 33 | 374 | 59 | 402 | |
| 2.48e-63 | 20 | 366 | 53 | 392 | |
| 1.72e-62 | 35 | 356 | 51 | 371 | |
| 2.71e-62 | 33 | 371 | 64 | 404 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.84e-62 | 41 | 361 | 29 | 368 | Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
|
| 3.60e-62 | 41 | 361 | 29 | 368 | The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_B The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_C The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3W6L_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
|
| 2.01e-61 | 41 | 361 | 62 | 401 | Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHM_B Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHM_C Crystal analysis of the complex structure, E342A-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii] |
|
| 2.01e-61 | 41 | 361 | 62 | 401 | Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHO_B Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii],3QHO_C Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii [Pyrococcus horikoshii] |
|
| 2.80e-61 | 41 | 361 | 62 | 401 | Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_A Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_B Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3],2ZUN_C Functional Analysis of Hyperthermophilic Endocellulase from the Archaeon Pyrococcus horikoshii [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.57e-58 | 44 | 356 | 58 | 369 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
|
| 9.60e-53 | 33 | 356 | 55 | 379 | Endoglucanase E1 OS=Acidothermus cellulolyticus (strain ATCC 43068 / DSM 8971 / 11B) OX=351607 GN=Acel_0614 PE=1 SV=1 |
|
| 2.77e-47 | 42 | 368 | 44 | 368 | Major extracellular endoglucanase OS=Xanthomonas campestris pv. campestris (strain ATCC 33913 / DSM 3586 / NCPPB 528 / LMG 568 / P 25) OX=190485 GN=engXCA PE=1 SV=2 |
|
| 2.55e-37 | 36 | 366 | 641 | 994 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
|
| 9.93e-34 | 42 | 366 | 62 | 441 | Endoglucanase G OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celG PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
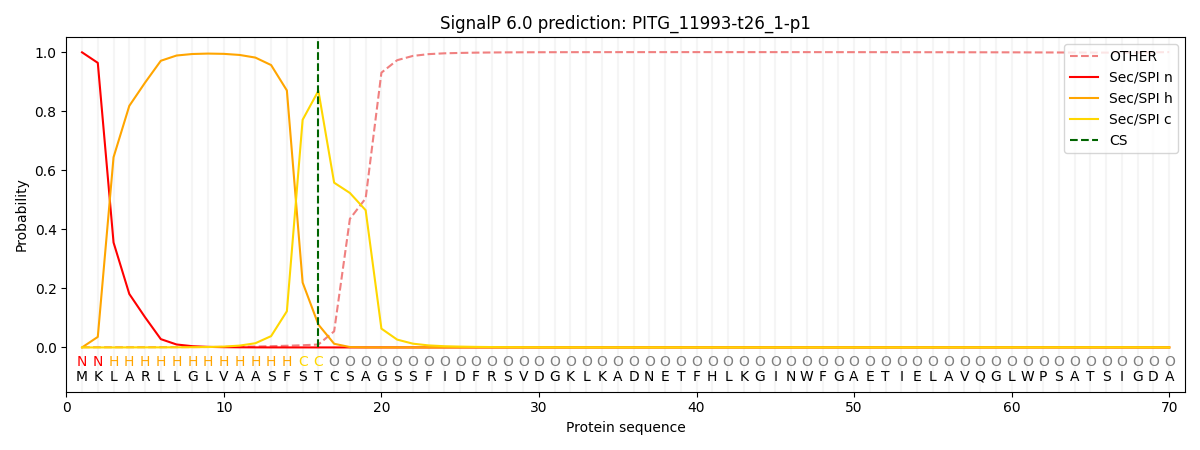
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001676 | 0.998306 | CS pos: 16-17. Pr: 0.8689 |

