You are browsing environment: FUNGIDB
CAZyme Information: PITG_09907-t26_1-p1
You are here: Home > Sequence: PITG_09907-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora infestans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans | |||||||||||
| CAZyme ID | PITG_09907-t26_1-p1 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | arabinogalactan endo-1,4-beta-galactosidase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.89:16 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 26 | 278 | 2.7e-68 | 0.7368421052631579 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 311610 | Glyco_hydro_53 | 4.44e-70 | 26 | 275 | 1 | 241 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| 226385 | GanB | 5.03e-47 | 18 | 271 | 32 | 283 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.30e-79 | 28 | 274 | 40 | 305 | |
| 2.98e-71 | 28 | 274 | 31 | 288 | |
| 5.04e-71 | 28 | 274 | 31 | 288 | |
| 8.08e-70 | 28 | 278 | 36 | 297 | |
| 1.33e-69 | 28 | 278 | 31 | 292 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.58e-63 | 28 | 276 | 6 | 252 | Crystal Structure Of Beta-1,4-galactanase From Aspergillus Aculeatus At 293k [Aspergillus aculeatus],1FOB_A Crystal Structure Of Beta-1,4-galactanase From Aspergillus Aculeatus At 100k [Aspergillus aculeatus],6Q3R_A ASPERGILLUS ACULEATUS GALACTANASE [Aspergillus aculeatus],6Q3R_B ASPERGILLUS ACULEATUS GALACTANASE [Aspergillus aculeatus] |
|
| 1.67e-62 | 33 | 276 | 10 | 251 | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJS_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_A Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_B Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_C Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus],1HJU_D Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Thermothelomyces thermophilus] |
|
| 1.05e-60 | 33 | 276 | 10 | 251 | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Humicola insolens] |
|
| 8.76e-58 | 20 | 275 | 15 | 268 | Emericilla nidulans endo-beta-1,4-galactanase [Aspergillus nidulans] |
|
| 9.88e-25 | 63 | 274 | 47 | 259 | Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482],6GPA_B Beta-1,4-galactanase from Bacteroides thetaiotaomicron with galactose [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.28e-66 | 20 | 276 | 14 | 268 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus tubingensis OX=5068 GN=galA PE=2 SV=1 |
|
| 3.60e-65 | 20 | 276 | 14 | 268 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=galA PE=3 SV=1 |
|
| 3.60e-65 | 20 | 276 | 14 | 268 | Arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus niger OX=5061 GN=galA PE=1 SV=1 |
|
| 5.69e-64 | 8 | 276 | 1 | 268 | Arabinogalactan endo-beta-1,4-galactanase OS=Aspergillus aculeatus OX=5053 GN=gal1 PE=1 SV=1 |
|
| 1.17e-62 | 28 | 275 | 22 | 265 | Probable arabinogalactan endo-beta-1,4-galactanase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=galA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
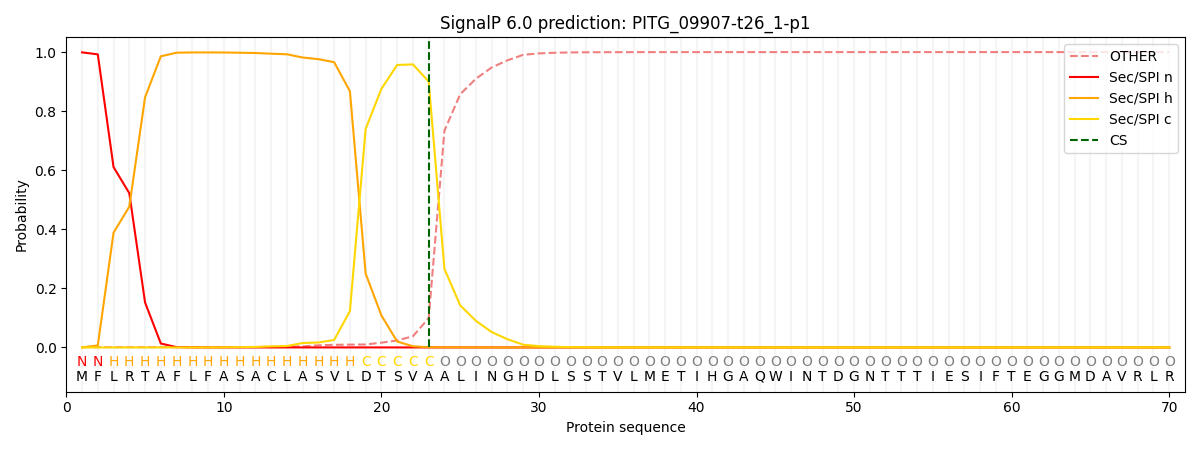
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.003534 | 0.996450 | CS pos: 23-24. Pr: 0.9003 |
