You are browsing environment: FUNGIDB
CAZyme Information: PITG_05097-t26_1-p1
Basic Information
help
| Species |
Phytophthora infestans
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans
|
| CAZyme ID |
PITG_05097-t26_1-p1
|
| CAZy Family |
GH12 |
| CAZyme Description |
glycoside hydrolase, putative
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 653 |
DS028124|CGC1 |
70347.99 |
4.6065 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PinfestansT30-4 |
19344 |
403677 |
1547 |
17797
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH78 |
88 |
595 |
1.7e-65 |
0.8630952380952381 |
PITG_05097-t26_1-p1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.89e-129 |
23 |
597 |
17 |
623 |
| 5.92e-127 |
23 |
597 |
22 |
632 |
| 5.92e-127 |
23 |
597 |
22 |
632 |
| 2.85e-126 |
23 |
597 |
21 |
627 |
| 2.92e-124 |
11 |
596 |
11 |
625 |
PITG_05097-t26_1-p1 has no PDB hit.
PITG_05097-t26_1-p1 has no Swissprot hit.
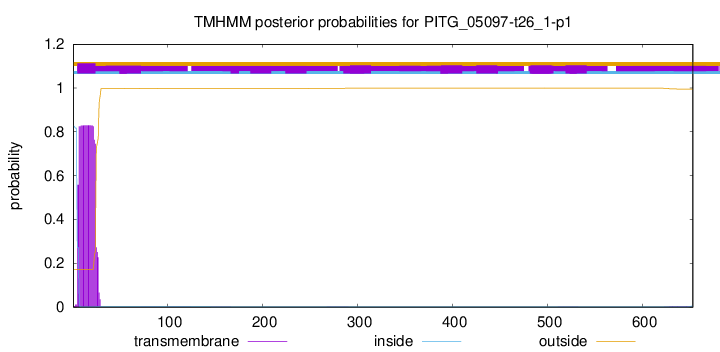
This protein is predicted as SP

| Other |
SP_Sec_SPI |
CS Position |
| 0.000357 |
0.999618 |
CS pos: 19-20. Pr: 0.9747 |