You are browsing environment: FUNGIDB
CAZyme Information: PITG_04949-t26_1-p1
You are here: Home > Sequence: PITG_04949-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora infestans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans | |||||||||||
| CAZyme ID | PITG_04949-t26_1-p1 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 11 | 260 | 6e-68 | 0.9591836734693877 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 237284 | PRK13108 | 4.72e-08 | 194 | 337 | 315 | 453 | prolipoprotein diacylglyceryl transferase; Reviewed |
| 236090 | PRK07764 | 7.68e-06 | 189 | 328 | 585 | 730 | DNA polymerase III subunits gamma and tau; Validated |
| 223039 | PHA03307 | 5.31e-05 | 198 | 337 | 257 | 392 | transcriptional regulator ICP4; Provisional |
| 380146 | MSCRAMM_SdrC | 2.18e-04 | 202 | 312 | 798 | 909 | MSCRAMM family adhesin SdrC. Features of this protein family include a YSIRK-type signal peptide at the N-terminus and a variable-length C-terminal region of Ser-Asp (SD) repeats followed by an LPXTG motif for surface immobilization by sortase. |
| 411231 | MSCRAMM_ClfA | 2.63e-04 | 202 | 307 | 561 | 668 | MSCRAMM family adhesin clumping factor ClfA. Clumping factor A is an MSCRAMM (Microbial Surface Components Recognizing Adhesive Matrix Molecules). It is heavily studied in Staphylococcus aureus both for its biological role in adhesion and for its potential for vaccination. Features of the sequence, but also of other MSCRAMM adhesins, include a long run of Ser-Asp dipeptide repeats and a C-terminal cell wall anchoring LPXTG motif. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.75e-179 | 1 | 337 | 1 | 337 | |
| 5.74e-154 | 1 | 224 | 1 | 224 | |
| 2.33e-153 | 1 | 224 | 1 | 224 | |
| 5.18e-139 | 1 | 212 | 1 | 212 | |
| 3.61e-111 | 1 | 219 | 1 | 220 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.74e-132 | 24 | 195 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
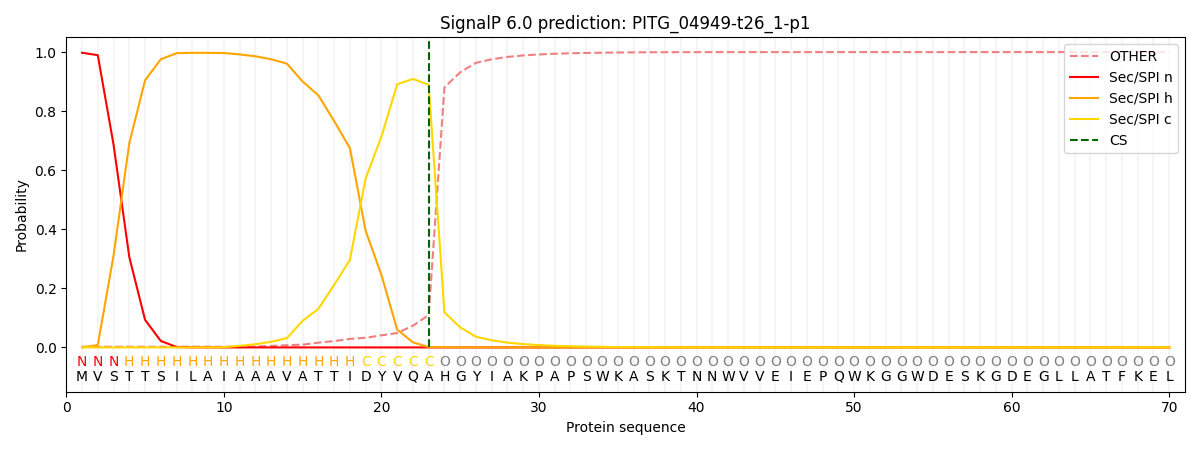
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.005254 | 0.994719 | CS pos: 23-24. Pr: 0.8893 |
