You are browsing environment: FUNGIDB
CAZyme Information: PITG_04615-t26_1-p1
You are here: Home > Sequence: PITG_04615-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
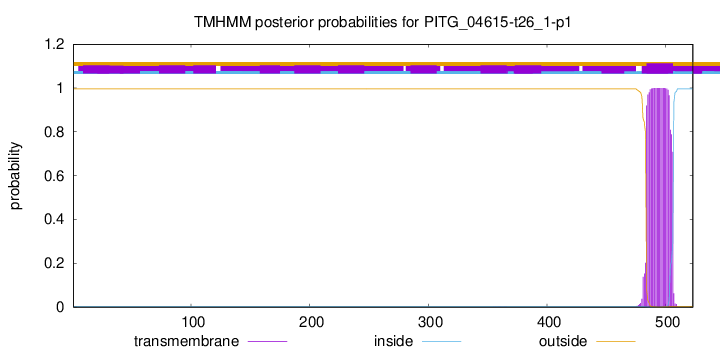
TMHMM annotations
Basic Information help
| Species | Phytophthora infestans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans | |||||||||||
| CAZyme ID | PITG_04615-t26_1-p1 | |||||||||||
| CAZy Family | GH1 | |||||||||||
| CAZyme Description | glucan 1,3-beta-glucosidase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 18 | 218 | 2.4e-109 | 0.7194244604316546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225344 | BglC | 1.42e-20 | 4 | 387 | 51 | 383 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| 197867 | X8 | 5.20e-14 | 404 | 469 | 13 | 79 | Possibly involved in carbohydrate binding. The X8 domain, which may be involved in carbohydrate binding, is found in an Olive pollen antigen as well as at the C terminus of family 17 glycosyl hydrolases. It contains 6 conserved cysteine residues which presumably form three disulfide bridges. |
| 400371 | X8 | 9.74e-12 | 404 | 461 | 13 | 76 | X8 domain. The X8 domain domain contains at least 6 conserved cysteine residues that presumably form three disulphide bridges. The domain is found in an Olive pollen allergen as well as at the C-terminus of several families of glycosyl hydrolases. This domain may be involved in carbohydrate binding. This domain is characteristic of GPI-anchored domains. |
| 396834 | Glyco_hydro_42 | 3.42e-04 | 22 | 180 | 6 | 138 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| 225019 | COG2108 | 0.001 | 13 | 152 | 105 | 212 | Uncharacterized conserved protein related to pyruvate formate-lyase activating enzyme [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 520 | 119 | 678 | |
| 1.06e-310 | 1 | 520 | 128 | 687 | |
| 1.15e-309 | 1 | 520 | 1 | 561 | |
| 3.16e-305 | 1 | 520 | 126 | 685 | |
| 5.34e-273 | 1 | 522 | 106 | 672 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.11e-41 | 1 | 391 | 43 | 384 | Exo-b-(1,3)-glucanase From Candida Albicans [Candida albicans] |
|
| 7.71e-41 | 1 | 391 | 43 | 384 | Exo-b-(1,3)-glucanase From Candida Albicans At 1.85 A Resolution [Candida albicans],1EQC_A Exo-b-(1,3)-glucanase From Candida Albicans In Complex With Castanospermine At 1.85 A [Candida albicans] |
|
| 8.70e-41 | 1 | 391 | 49 | 390 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
|
| 8.70e-41 | 1 | 391 | 49 | 390 | Chain A, Hypothetical protein XOG1 [Candida albicans] |
|
| 8.70e-41 | 1 | 391 | 49 | 390 | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A [Candida albicans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.94e-41 | 1 | 391 | 57 | 396 | Probable glucan 1,3-beta-glucosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=exgA PE=3 SV=2 |
|
| 3.52e-40 | 1 | 391 | 87 | 428 | Glucan 1,3-beta-glucosidase OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=XOG1 PE=1 SV=5 |
|
| 5.10e-40 | 1 | 416 | 74 | 479 | Glucan 1,3-beta-glucosidase 1 OS=Wickerhamomyces anomalus OX=4927 GN=EXG1 PE=3 SV=1 |
|
| 3.29e-37 | 1 | 387 | 67 | 405 | Glucan 1,3-beta-glucosidase OS=Blumeria graminis OX=34373 PE=3 SV=1 |
|
| 2.02e-36 | 1 | 391 | 83 | 430 | Glucan 1,3-beta-glucosidase OS=Lachancea kluyveri (strain ATCC 58438 / CBS 3082 / BCRC 21498 / NBRC 1685 / JCM 7257 / NCYC 543 / NRRL Y-12651) OX=226302 GN=EXG1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
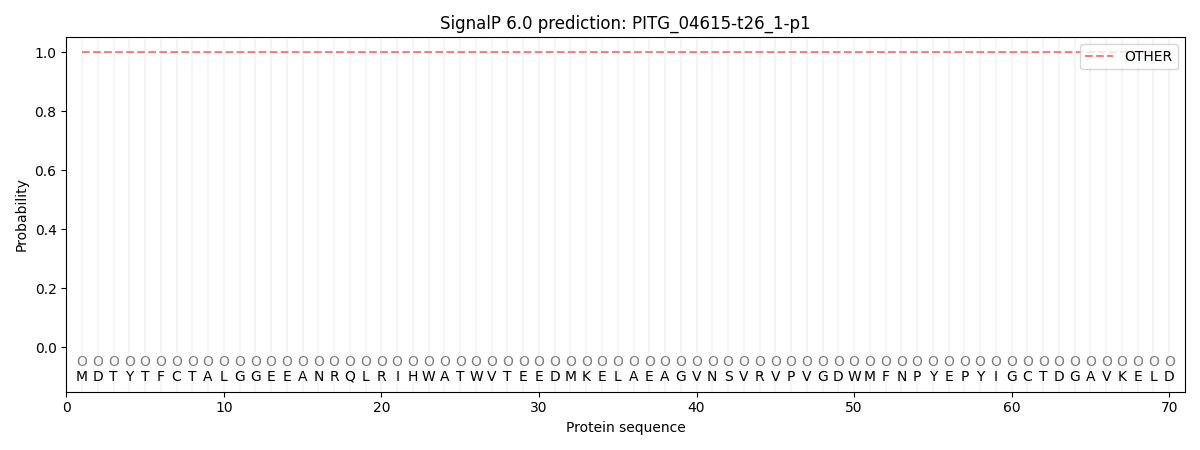
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000049 | 0.000000 |