You are browsing environment: FUNGIDB
CAZyme Information: PITG_04158-t26_1-p1
Basic Information
help
| Species |
Phytophthora infestans
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans
|
| CAZyme ID |
PITG_04158-t26_1-p1
|
| CAZy Family |
GH1 |
| CAZyme Description |
glycoside hydrolase, putative
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 298 |
|
33361.38 |
6.7400 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PinfestansT30-4 |
19344 |
403677 |
1547 |
17797
|
|
| Gene Location |
No EC number prediction in PITG_04158-t26_1-p1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH17 |
26 |
280 |
1.1e-19 |
0.977491961414791 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227625
|
Scw11 |
7.63e-21 |
45 |
276 |
68 |
304 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033
|
Glyco_hydro_17 |
3.12e-04 |
175 |
283 |
173 |
307 |
Glycosyl hydrolases family 17. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 4.21e-144 |
7 |
284 |
10 |
288 |
| 9.47e-98 |
19 |
284 |
567 |
834 |
| 7.39e-77 |
10 |
287 |
13 |
291 |
| 9.70e-53 |
10 |
196 |
13 |
200 |
| 1.66e-45 |
26 |
285 |
22 |
283 |
PITG_04158-t26_1-p1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 2.29e-12 |
109 |
260 |
120 |
287 |
Probable glucan endo-1,3-beta-glucosidase eglC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglC PE=1 SV=1 |
This protein is predicted as SP
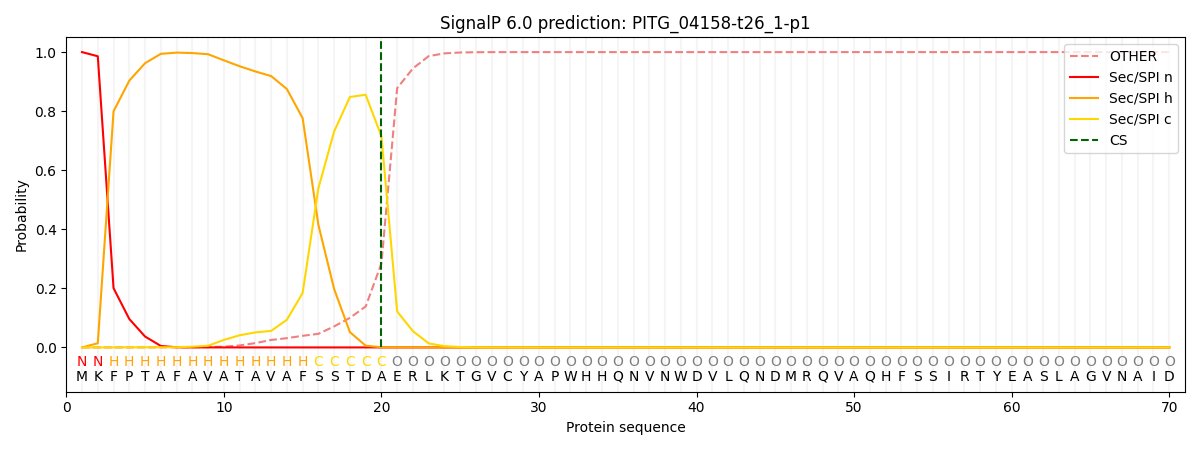
| Other |
SP_Sec_SPI |
CS Position |
| 0.000635 |
0.999349 |
CS pos: 20-21. Pr: 0.7152 |
There is no transmembrane helices in PITG_04158-t26_1-p1.

