You are browsing environment: FUNGIDB
CAZyme Information: PITG_03979-t26_1-p1
You are here: Home > Sequence: PITG_03979-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora infestans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans | |||||||||||
| CAZyme ID | PITG_03979-t26_1-p1 | |||||||||||
| CAZy Family | CE8|CE8 | |||||||||||
| CAZyme Description | conserved hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 95 | 336 | 5.9e-18 | 0.8938906752411575 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.52e-23 | 56 | 333 | 45 | 305 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 226973 | COG4625 | 0.004 | 102 | 294 | 117 | 313 | Uncharacterized conserved protein, contains a C-terminal beta-barrel porin domain [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 5.15e-144 | 121 | 380 | 16 | 279 | |
| 3.46e-110 | 58 | 343 | 94 | 381 | |
| 5.05e-100 | 53 | 343 | 299 | 591 | |
| 1.70e-94 | 53 | 332 | 20 | 304 | |
| 1.31e-91 | 56 | 304 | 309 | 559 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.16e-09 | 84 | 336 | 44 | 296 | Glucan 1,3-beta-glucosidase ARB_02797 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02797 PE=1 SV=1 |
|
| 5.73e-07 | 58 | 295 | 304 | 536 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 1.78e-06 | 58 | 332 | 310 | 565 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=btgE PE=3 SV=2 |
|
| 1.78e-06 | 58 | 332 | 310 | 565 | Probable beta-glucosidase btgE OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=btgE PE=3 SV=2 |
|
| 3.22e-06 | 58 | 327 | 348 | 596 | Probable beta-glucosidase btgE OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
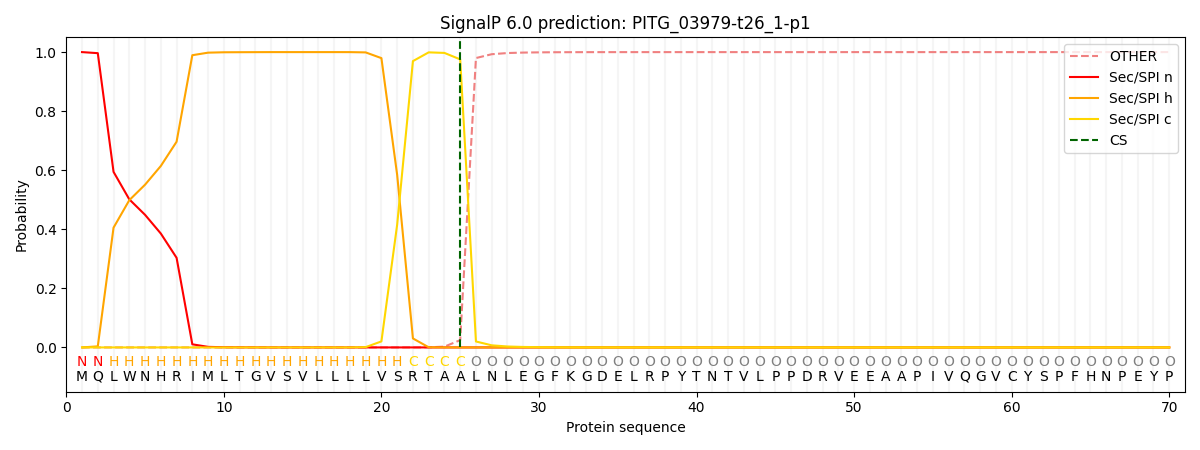
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000216 | 0.999759 | CS pos: 25-26. Pr: 0.9754 |
