You are browsing environment: FUNGIDB
CAZyme Information: PITG_01398-t26_1-p1
You are here: Home > Sequence: PITG_01398-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
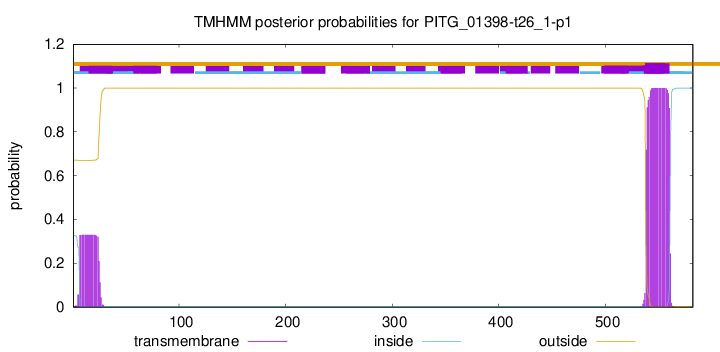
TMHMM annotations
Basic Information help
| Species | Phytophthora infestans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora infestans | |||||||||||
| CAZyme ID | PITG_01398-t26_1-p1 | |||||||||||
| CAZy Family | AA17 | |||||||||||
| CAZyme Description | beta-glucosidase, putative | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.21:2 | 3.2.1.23:1 | 3.2.1.38:1 | 3.2.1.-:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH1 | 28 | 510 | 7.6e-145 | 0.986013986013986 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395176 | Glyco_hydro_1 | 4.01e-152 | 30 | 511 | 5 | 451 | Glycosyl hydrolase family 1. |
| 274539 | BGL | 9.43e-140 | 31 | 504 | 1 | 426 | beta-galactosidase. |
| 225343 | BglB | 9.32e-127 | 30 | 512 | 4 | 454 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| 215435 | PLN02814 | 1.77e-97 | 10 | 504 | 8 | 476 | beta-glucosidase |
| 215455 | PLN02849 | 3.43e-96 | 1 | 504 | 1 | 476 | beta-glucosidase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 582 | 1 | 586 | |
| 0.0 | 23 | 582 | 6 | 562 | |
| 5.90e-217 | 14 | 512 | 7 | 505 | |
| 8.61e-150 | 78 | 514 | 5 | 419 | |
| 1.07e-113 | 30 | 515 | 4 | 460 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.13e-100 | 30 | 503 | 22 | 506 | Crystal structure of native Raucaffricine glucosidase [Rauvolfia serpentina],4ATD_B Crystal structure of native Raucaffricine glucosidase [Rauvolfia serpentina],4ATL_A Crystal structure of Raucaffricine glucosidase in complex with Glucose [Rauvolfia serpentina],4ATL_B Crystal structure of Raucaffricine glucosidase in complex with Glucose [Rauvolfia serpentina] |
|
| 8.45e-100 | 17 | 506 | 26 | 503 | Crystal structure of beta-primeverosidase [Camellia sinensis],3WQ4_B Crystal structure of beta-primeverosidase [Camellia sinensis],3WQ5_A beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, natural aglycone derivative [Camellia sinensis],3WQ5_B beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, natural aglycone derivative [Camellia sinensis],3WQ6_A beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, artificial aglycone derivative [Camellia sinensis],3WQ6_B beta-Primeverosidase in complex with disaccharide substrate-analog N-beta-primeverosylamidine, artificial aglycone derivative [Camellia sinensis] |
|
| 1.53e-99 | 30 | 503 | 22 | 506 | Crystal of Raucaffricine Glucosidase in complex with inhibitor [Rauvolfia serpentina],3ZJ6_B Crystal of Raucaffricine Glucosidase in complex with inhibitor [Rauvolfia serpentina],4A3Y_A Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway [Rauvolfia serpentina],4A3Y_B Crystal structure of Raucaffricine glucosidase from ajmaline biosynthesis pathway [Rauvolfia serpentina] |
|
| 1.99e-99 | 30 | 503 | 22 | 506 | Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity [Rauvolfia serpentina],3U57_B Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity [Rauvolfia serpentina],3U5U_A Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity [Rauvolfia serpentina],3U5U_B Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity [Rauvolfia serpentina],3U5Y_A Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity [Rauvolfia serpentina],3U5Y_B Structures of Alkaloid Biosynthetic Glucosidases Decode Substrate Specificity [Rauvolfia serpentina],4EK7_A High speed X-ray analysis of plant enzymes at room temperature [Rauvolfia serpentina],4EK7_B High speed X-ray analysis of plant enzymes at room temperature [Rauvolfia serpentina] |
|
| 4.06e-99 | 30 | 507 | 13 | 459 | Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8],4GXP_B Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8],4GXP_C Chimeric Family 1 beta-glucosidase made with non-contiguous SCHEMA [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.42e-107 | 21 | 505 | 28 | 506 | Beta-glucosidase 32 OS=Arabidopsis thaliana OX=3702 GN=BGLU32 PE=2 SV=2 |
|
| 6.49e-105 | 26 | 524 | 27 | 516 | Beta-glucosidase 30 OS=Arabidopsis thaliana OX=3702 GN=BGLU30 PE=2 SV=1 |
|
| 5.80e-104 | 26 | 506 | 30 | 502 | Beta-glucosidase 28 OS=Arabidopsis thaliana OX=3702 GN=BGLU28 PE=2 SV=1 |
|
| 2.45e-103 | 30 | 506 | 19 | 490 | Beta-glucosidase 26, peroxisomal OS=Arabidopsis thaliana OX=3702 GN=BGLU26 PE=1 SV=1 |
|
| 1.29e-102 | 21 | 505 | 28 | 506 | Beta-glucosidase 31 OS=Arabidopsis thaliana OX=3702 GN=BGLU31 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
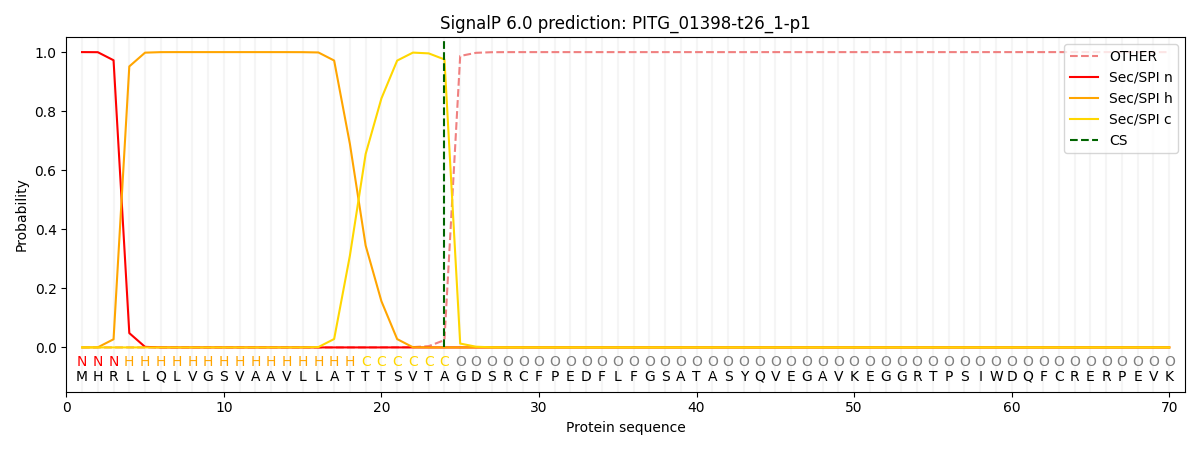
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000272 | 0.999700 | CS pos: 24-25. Pr: 0.9752 |