You are browsing environment: FUNGIDB
CAZyme Information: PHYSODRAFT_557002-t26_1-p1
You are here: Home > Sequence: PHYSODRAFT_557002-t26_1-p1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora sojae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora sojae | |||||||||||
| CAZyme ID | PHYSODRAFT_557002-t26_1-p1 | |||||||||||
| CAZy Family | PL3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.99.18:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 22 | 552 | 8.4e-110 | 0.9726277372262774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 2.86e-36 | 28 | 558 | 8 | 539 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 5.21e-21 | 28 | 350 | 6 | 309 | choline dehydrogenase; Validated |
| 366272 | GMC_oxred_N | 2.86e-16 | 110 | 329 | 19 | 215 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 238533 | APPLE_Factor_XI_like | 2.15e-12 | 641 | 709 | 3 | 70 | Subfamily of PAN/APPLE-like domains; present in plasma prekallikrein/coagulation factor XI, microneme antigen proteins, and a few prokaryotic proteins. PAN/APPLE domains fulfill diverse biological functions by mediating protein-protein or protein-carbohydrate interactions. |
| 238533 | APPLE_Factor_XI_like | 3.30e-12 | 719 | 784 | 7 | 71 | Subfamily of PAN/APPLE-like domains; present in plasma prekallikrein/coagulation factor XI, microneme antigen proteins, and a few prokaryotic proteins. PAN/APPLE domains fulfill diverse biological functions by mediating protein-protein or protein-carbohydrate interactions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.40e-266 | 9 | 635 | 9 | 637 | |
| 6.71e-217 | 28 | 785 | 24 | 811 | |
| 2.56e-195 | 7 | 782 | 3 | 807 | |
| 3.34e-148 | 13 | 534 | 7 | 526 | |
| 4.16e-141 | 7 | 556 | 3 | 553 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.91e-45 | 24 | 552 | 4 | 539 | Chain A, cellobiose dehydrogenase [Phanerodontia chrysosporium],1KDG_B Chain B, cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 1.01e-44 | 28 | 552 | 3 | 534 | Chain A, Cellobiose dehydrogenase [Phanerodontia chrysosporium],1NAA_B Chain B, Cellobiose dehydrogenase [Phanerodontia chrysosporium] |
|
| 1.36e-34 | 28 | 552 | 8 | 542 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI5_A Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 4.91e-34 | 28 | 552 | 230 | 764 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
|
| 3.04e-30 | 28 | 552 | 230 | 763 | Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A],4QI7_B Cellobiose dehydrogenase from Neurospora crassa, NcCDH [Neurospora crassa OR74A] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.78e-43 | 24 | 552 | 231 | 766 | Cellobiose dehydrogenase OS=Phanerodontia chrysosporium OX=2822231 GN=CDH-1 PE=1 SV=1 |
|
| 1.17e-19 | 24 | 554 | 3 | 535 | 4-pyridoxate dehydrogenase OS=Mesorhizobium japonicum (strain LMG 29417 / CECT 9101 / MAFF 303099) OX=266835 GN=padh1 PE=1 SV=1 |
|
| 9.40e-15 | 26 | 528 | 3 | 505 | Oxygen-dependent choline dehydrogenase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=betA PE=3 SV=1 |
|
| 1.21e-14 | 29 | 552 | 4 | 525 | Oxygen-dependent choline dehydrogenase OS=Brucella anthropi (strain ATCC 49188 / DSM 6882 / CCUG 24695 / JCM 21032 / LMG 3331 / NBRC 15819 / NCTC 12168 / Alc 37) OX=439375 GN=betA PE=3 SV=1 |
|
| 1.59e-14 | 29 | 552 | 4 | 525 | Oxygen-dependent choline dehydrogenase OS=Brucella ovis (strain ATCC 25840 / 63/290 / NCTC 10512) OX=444178 GN=betA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
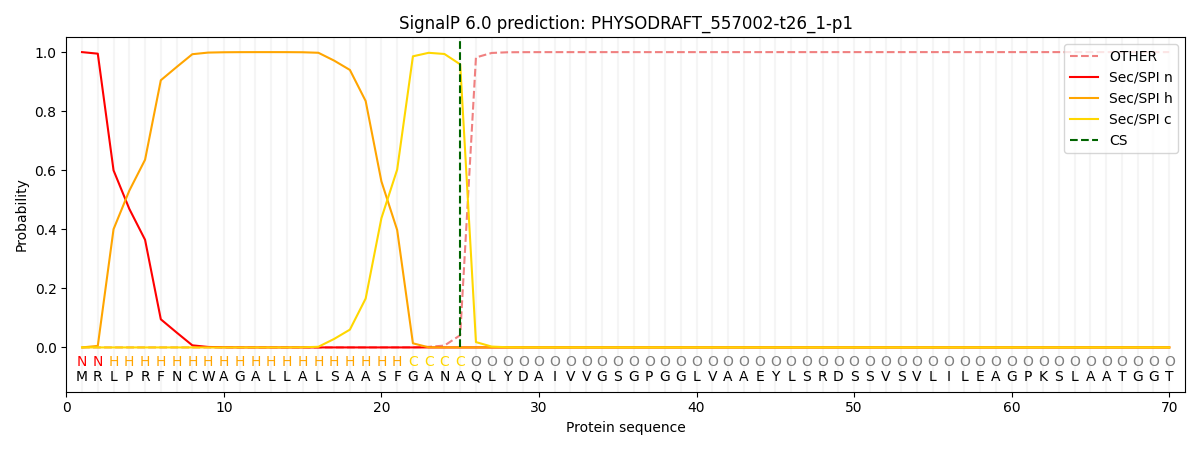
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000576 | 0.999376 | CS pos: 25-26. Pr: 0.9586 |
